| wiki | search |
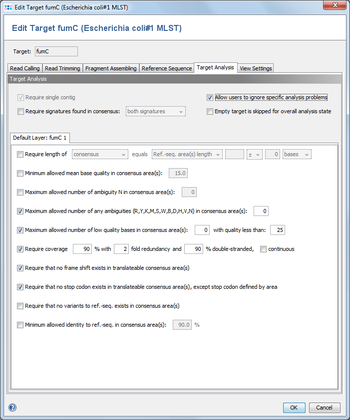
Allows to define different requirements that must be fulfilled by a target to reach the green smiley ![]() . If they are not all fulfilled the target gets the yellow icon
. If they are not all fulfilled the target gets the yellow icon ![]() (the red icon
(the red icon ![]() means that no contigs can be assembled).
means that no contigs can be assembled).
Except for the first three requirements, all others refer to the first contig if a target has several contigs.
Layer Independent Options
- Require Single Contig
- Always set and cannot be disabled. All further analysis is done on the first contig only. Therefore the analysis fails if the reads were assembled into multiple contigs.
- Allow users to ignore specific analysis problems
- If this option is checked, an Ignore button is added to the analysis problem table in sequence view.
- Empty target is skipped for overall analysis state
- By default, targets without sequence will set the analysis state of the whole task entry to 'bad'. If this option is set, any targets without sequence will be ignored during analysis and will have no effect on the analysis state of the task entry.
- Require signatures found in consensus
- Defines that the 3` signature, the 5`signature or both must be found in the consensus.
Layer Dependent Options
- Require consensus (areas) length
- Defines that the trimmed consensus, or all bases of the consensus that are covered by a ref.-seq. area, must have a specific length, or the same length as the reference sequence in all areas of this layer. A tolerance range for the length comparison can be set in bases or codons.
- Minimum allowed mean base quality in consensus area(s)
- Defines the allowed minimum mean base quality, calculated for all bases in the consensus areas of this layer. This feature is only available for Sanger sequencing data.
- Maximum allowed number of low quality bases in consensus area(s)
- Defines the allowed maximum number of low quality bases in the consensus areas of this layer. The threshold for a low quality can be specified. This feature is only available for Sanger sequencing data.
- Maximum allowed number of ambiguities N in consensus area(s)
- Defines the allowed maximum number of the ambiguity N in the consensus areas of this layer
- Maximum allowed number of any ambiguities in consensus area(s)
- Defines the allowed maximum number of any ambiguities in the consensus areas of this layer
- Require coverage (Sanger Sequencing)
- Defines the coverage that is demanded for the bases in the consensus areas of this layer, and optionally if both reading directions (forward and reverse) should be covered in the reads. The coverage is defined by percentage for to the base count in the consensus areas. To handle badly covered ends of the contig, it can be defined that the uncovered positions (if demanded coverage <100%) may only appear on the ends of the contig. This feature is only available for Sanger sequencing data.
- Require coverage (Whole Genome Sequencing)
- Defines the coverage that is demanded for the bases in the consensus areas of this layer in any reading direction. This feature is only available for whole genome sequencing data.
- Require that no frame shift (non-triplet gaps) exists in translatable consensus area(s)
- Defines that that only in-frame gaps (length dividable by 3) may appear in the alignment.
- Require that no stop-codon exists in translatable consensus area(s), except defined by area
- Defines that the translation of the consensus areas according to the layer may not contain a stop codon (beside the last codon if the last area is defined with "ends with stop")
- Require that no unknown variant exists
- Demand that all differences between the consensus and the ref.-seq. in the areas of this layer must be defined as known variants. Known variants can be defined in the Reference Sequence Parameters.
- Minimum allowed identity to ref.-seq. in consensus area(s)
- Identity is calculated as the rational difference between consensus and reference sequence in an alignment. Differences are gaps and mismatches. Ambiguities never match to bases, but to the same ambiguity.