Ridom SeqSphere+ allows to submit raw read data (FASTQ) together with epidemiological data of a Sample to the European Nucleotide Archive (ENA).
As for all public submission in SeqSphere+ the Submission Anonymization Filter is shown as first step.
In the next step the raw reads are selected for the submission. If the Sample was created from raw reads in pipeline mode, the FASTQs files are associated to the Sample and are automatically filled in and the dialog can be confirmed.
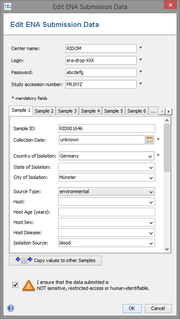
Next the submission dialog window opens, and shows the ENA account specifications and the submission data for each Sample. Mandatory fields are marked by *.
The ENA Account specifications consists of
- Center name
- Login
- Password
- Study accession number
The first 3 values are stored per SeqSphere+ user in the database and can be edited with the menu item Options | User Settings. The study accession number must be entered manually for each submission. The study must already be existing.
New studies and accounts can be created at ENA Webin.
Initial editor values are taken from the Sample's data fields and from the Procedure Details, anonymized according to the settings in the Submission Anonymization Filter. The Copy values to other editors button can be used to copy data from the editor in the currently selected tab to all other editors. Make sure the box next to "I ensure that the data submitted is ..." is selected, then click OK to start the submission to ENA.
If the Samples use a Database Scheme that contains the field EBI/NCBI Accession(s), the ENA run accession numbers are appended to this field.
![]() Hint: A Source Type other than "unknown" must be given if a value for Isolation Source is specified. If Source Type is "clinical/host-associated" a value for Host must be given if a value for Isolation Source is specified. Host, Host Age, Host Sex, and Host Disease can be only submitted if Source Type is "clinical/host-associated" and a Host is given. Host Disease is submitted as "host_disease_stage" if it is "healthy". Other values are ignored and not submitted.
Hint: A Source Type other than "unknown" must be given if a value for Isolation Source is specified. If Source Type is "clinical/host-associated" a value for Host must be given if a value for Isolation Source is specified. Host, Host Age, Host Sex, and Host Disease can be only submitted if Source Type is "clinical/host-associated" and a Host is given. Host Disease is submitted as "host_disease_stage" if it is "healthy". Other values are ignored and not submitted.
![]() Warning: Make sure the entered data is correct because it might be difficult or impossible to remove the data from ENA once a Sample is submitted!
Warning: Make sure the entered data is correct because it might be difficult or impossible to remove the data from ENA once a Sample is submitted!