Contents
Introduction
Here we compare as of April 2024 the contiguity and accuracy results of the Oxford Nanopore Technologies (ONT) Native (NBK), Rapid (RBK), and Rapid-PCR Barcoding Kits (RPBK).
Methods
Seven culture collections strains that have finished NCBI RefSeq genomes were re-sequenced with Illumina DNA Prep, PacBio HiFi v3, and ONT NBK, RBK, and RPBK (with NEB LongAmp Taq) library preparations and then loading on a MiSeq, Sequel IIe, or MinION R10.4.1 flowcell, respectively. These RefSeq isolates spanned a GC range from 32.1-73.1%. Additionally, a couple of isolates from a ring-trial organized by Gabriel Wagner (Univ. Graz, Austria) with methylation profiles causing strand-specific errors with the ONT technology were sequenced with PacBio and ONT. DNA was extracted with the Zymo Biomics DNA Miniprep kit (Zymo Research; Freiburg, Germany). Basecalling was done with Dorado (v0.5.3) with model dna_r10.4.1_e8.2_400bps_sup@v4.3.0 either in simplex or duplex mode. Reads were trimmed with chopper (v0.7.0; with quality 10 and minimum length 500) and then de novo assembled with Flye (v2.9.3; with option --nano-hq). Consensus sequences were polished with Medaka (v1.11.3) either with r1041_ e82_400bps_sup_v4.3.0 (‘Dorado’) or r103_min_high_g360 (‘Guppy’) model. Plasmid reconstruction was done with MOB-recon. Seed-only cgMLST task templates that were defined from the downloaded NCBI genomes or the PacBio genomes of the same strains were used with SeqSphere+. Whole genomes were compared and analyzed with Quast (v5.2.0). Please note that the rates of InDels and substitutions between the assemblies and NCBI genomes (smaller values are better) reported by Quast will never be zero due to sequencing errors in the finished NCBI genomes and/or micro-evolutionary changes of the re-sequenced culture collection strains. NanoComp (v1.23.1) was used to compare sample specific quality metrics on read level. BRIG (v0.95) generated the genome comparison figure.
NCBI RefSeq Strains
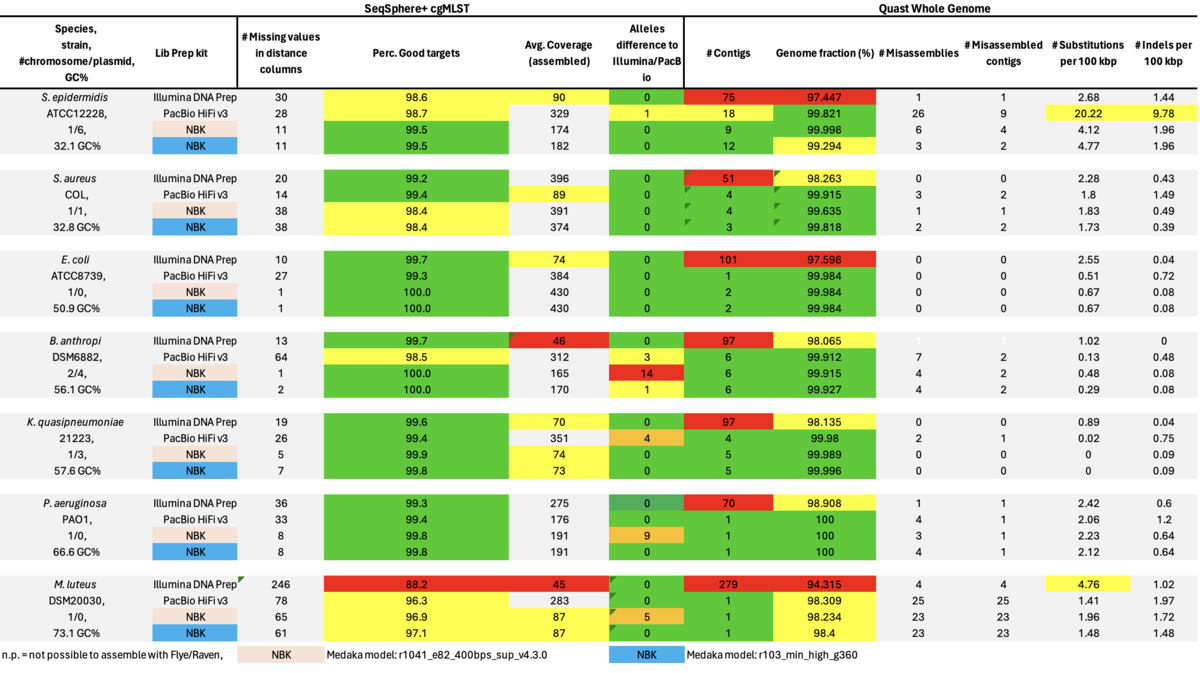
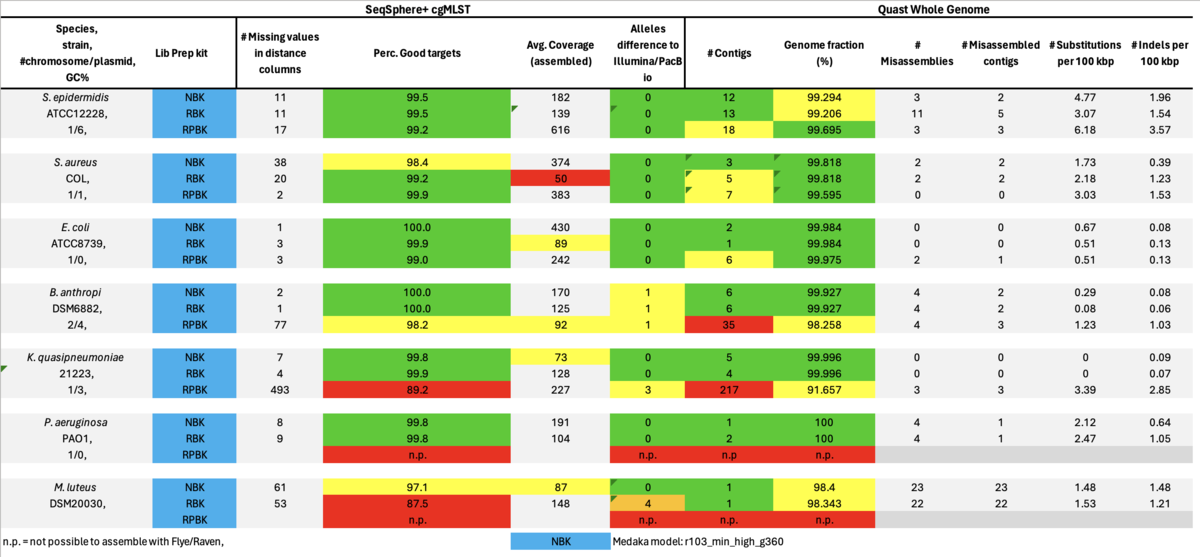
As can be seen it was not possible to achieve assemblies with the RPBK data for the two high GC content isolates, i.e., P. aeruginosa (66.6%) and M. luteus (73.1% GC). Therefore, we also took a closer look at some missing cgMLST target blocks of the RPBK K. quasipneumoniae data which had on average a GC content of 57.6%. The seven blocks inspected ranged in size between 1.5 and 13.3 kb and had a GC content ranging from 62.1 to 65.8%.
Table: Illumina DNA Prep, PacBio HiFi, and ONT NBK with ‘Dorado’ and ‘Guppy’ models for Medaka polishing.
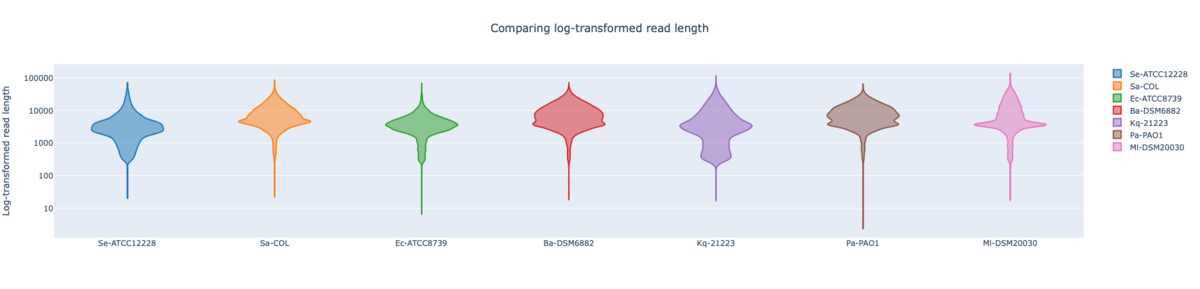
Figure: NBK log-transformed read lengths of the seven RefSeq samples with average read length N50 9.96 kb (mean read quality 18.3).
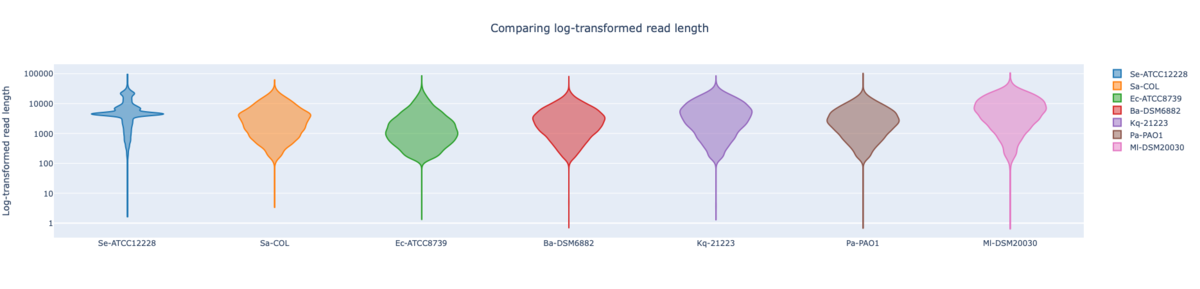
Figure: RBK log-transformed read lengths of the seven RefSeq samples with average read length N50 8.62 kb (mean read quality 16.7).
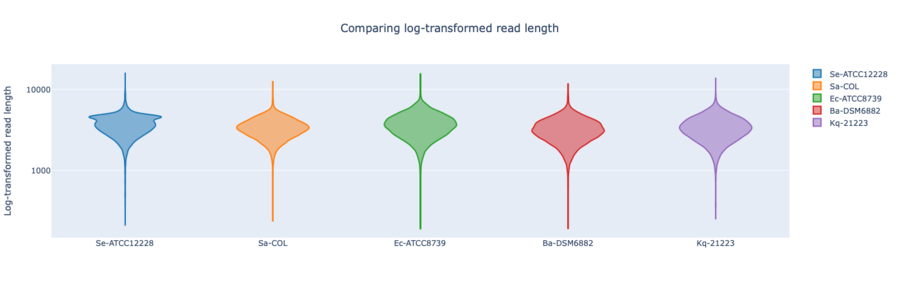
Figure: RPBK log-transformed read lengths of the seven RefSeq samples with average read length N50 3.69 kb (mean read quality 17).
Table: ONT NBK, RBK, and RPBK all with ‘Guppy’ model for Medaka polishing.
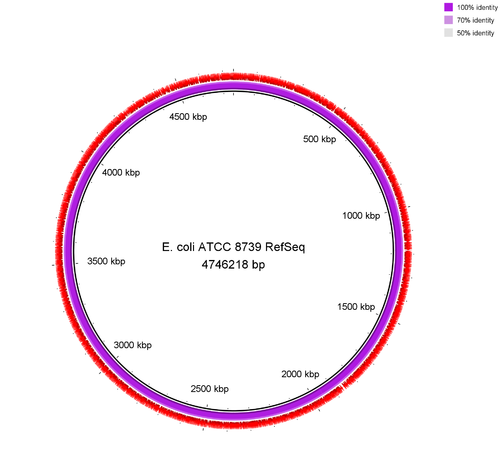
Figure: Genome comparison of E. coli ATTC 83739 re-sequenced with ONT-RBK against the NCBI RefSeq (GCF_000019385.1) genome of the very same isolate. Flye generated a circular chromosome and according to Quast analysis there was no single misassembly.
In summary, for Illumina DNA Prep, PacBio, and ONT NBK a bias at very high GC% was observed (here at 73.1% GC). ONT NBK whole genome contiguity and accuracy was on similar level as for the PacBio data. The ‘Guppy’ medaka model further improved the cgMLST accuracy in comparison to the ‘Dorado’ model. Read length was largest for NBK followed by RBK and then RPBK. However, quite a species-specific variation in read length was noted. Read quality was slightly better for NBK. There was hardly any difference in the contiguity between NBK and RBK. Contiguity of RPBK was not so good but still substantially better than for Illumina. A serious GC bias was noted for RPBK starting at around 60%. However, a more processive Taq polymerase and/or longer elongation times might solve the issue.
Strains with Known Modification-mediated Errors
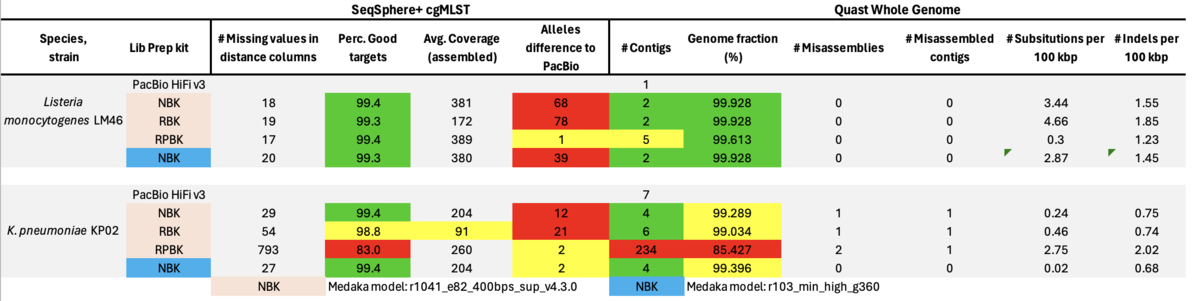
ONT published in April 2023 a note informing customers of possible issues with strand-specific errors due to methylation. In a preprint by Mara Lohde et al. the authors found in over 40% of 264 screened isolates of in total 32 different species more than 50 problematic positions in each of the NBK genomes (involving 25 of the 32 species; [bioRxiv]). The authors recommended using RPBK to get rid of the issues but evaluated only eight K. pneumoniae isolates with this kit. Finally, Lohde et al. suggested utilizing NBK and RPBK sequencing kits for library preparation and pooling their libraries in approximately 30/70 ratio prior to flowcell loading. In the following, only results from 2 exemplary strains with same issues are shown as they summarize all our observations.
Table: PacBio HiFi (that was used for comparison as no NCBI RefSeqs were available), ONT NBK, RBK, and RPBK all with ‘Dorado’ and ONT NBK with ‘Guppy’ model for Medaka polishing.
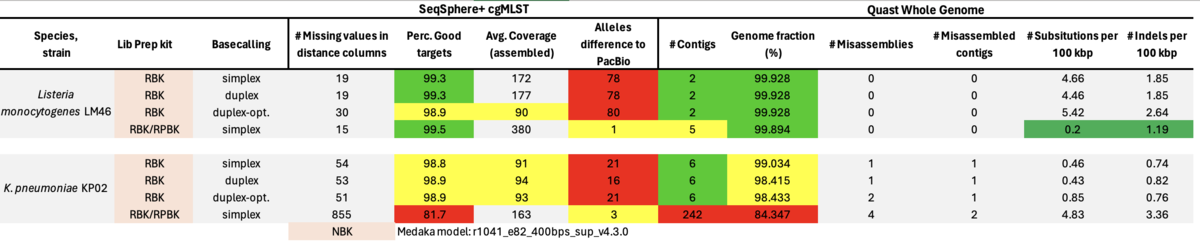
In another preprint by Fabian Landman et al. the authors made a rather large study involving 356 MDROs using RBK with Dorado duplex mode basecalling [medRxiv]. Comparing Illumina with RBK results the authors found only one to nine wgMLST allele differences (just for P. aeruginosa up to 27 alle differences). Following, we evaluated this recommendation (with on average about 2% duplex reads) and also made a run with duplex optimized conditions with pore occupancies of only about 50% (resulting in about 20% duplex reads) and therefore only half of the possible flowcell throughput.
Table: ONT RBK with simplex, ONT RBK and ONT RBK optimized with duplex, and ONT RBK/RPBK (30/70 ratio) with simplex Dorado basecalling. All data were polished with the ‘Dorado’ model for Medaka polishing.
Finally, we did some preliminary evaluation with the Modpolish tool that corrects modification-mediated errors in ONT sequencing by nucleotide demodification and reference-based correction. The tool was able for example to correct all but one error in our ‘problematic’ Listeria isolate from above. However, it is very compute-intensive and we observed also instances where the software even introduced errors in previously correct genomes.
In summary, the Medaka ‘Guppy’ model reduced but did not remove all cgMLST errors of ‘problematic’ strains. The amplification step with RPBK eliminated (nearly) all errors due to methylation. Data basecalling with Dorado in duplex mode had almost no beneficial effect. The same was true for duplex-optimized runs. With a ratio of about 30/70% pooled RBK/RPBK libraries all errors were eliminated, but contiguity seemed to be slightly worse as with RBK-only. Furthermore, high GC RPBK gaps were not bridged by the RBK reads. Modpolish not only relies on others to produce high-quality reference genomes but is also very compute-intensive and did not always produce correct genome sequences.
Conclusion
For a reliable fine scale analysis (e.g., SNV or cgMLST genotyping) with ONT the RPBK library preparation should be used. For structural analysis (e.g., plasmid characterization) ONT RBK-only – as with NBK especially smaller plasmids are not so well recovered - assemblies suffices. However, in case of high GC issues with RPBK, or if best contiguity and accuracy is aimed for, then assemblies with ONT RBK and Illumina reads for polishing are still the best solution. Similar conclusions were obtained by a recent publication by Nicole Lerminiaux et al. [PubMed 38354391].