Contents
Preparation
Before whole genome sequence data can be imported a Project with at least one Task Template for whole genome sequencing data must be existing in the database.
![]() Important: When working with read files (FASTQ) the Pipeline Mode should be used instead.
Important: When working with read files (FASTQ) the Pipeline Mode should be used instead.
Choose Input WGS Data
Use the menu function ![]() File | Process Assembled Genome Data to open a dialog window.
File | Process Assembled Genome Data to open a dialog window.
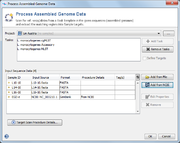
A Project and at least one Task Template must be selected. If a single Task Template is selected, the process can be limited to specific targets using the Define Targets checkbox.
In the Input Sequence Data section the files with whole genome sequence data can be selected. It is possible to either
- to add files using the
 Add from file-button, or
Add from file-button, or
- to add sequences from NCBI GenBank using the
 Add from NCBI button.
Add from NCBI button.
Allowed input file formats are FASTA, GenBank, SAM/BAM and ACE-files. If SAM/BAM files do not contain a reference sequence, a dialog windows opens that allows to specify a FASTA-file with the reference sequence. The sequence names in the FASTA-file must match the names in the SAM/BAM file.
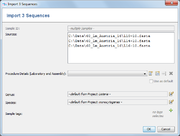
If on or more files are added file to the list, a dialog with details are shown. This allows to view and edit:
- Sample ID (if only one one file was chosen)
- Laboratory and Assembly Procedure Details
- Genus (default is taken from Project, or for GenBank data from file)
- Species (default is taken from Project, or for GenBank data from file)
- Sample tags
- Assembly File Management
After confirming, the files are added with the as samples to the preview table. The ![]() icon is shown, if the Sample ID does not exist in the project yet. If the Sample ID is already existing in the project, the icon of the existing sample is shown, and the right-click menu offers functions to open the existing sample(s) or to delete it directly. If the dialog is confirmed with Sample IDs of existing samples, a warning will be shown. The targets of the existing samples will be overwritten, if overwriting was enabled in the General Settings.
icon is shown, if the Sample ID does not exist in the project yet. If the Sample ID is already existing in the project, the icon of the existing sample is shown, and the right-click menu offers functions to open the existing sample(s) or to delete it directly. If the dialog is confirmed with Sample IDs of existing samples, a warning will be shown. The targets of the existing samples will be overwritten, if overwriting was enabled in the General Settings.
Press the button Target Scan Procedure Details... to show the parameters:
- General Options
- Ignore contigs shorter than: This setting is used to improve the speed by ignoring contigs in the input file that are shorter than a minimum length (default: 200bp).
- Perform auto-correction for homopolymer errors for IonTorrent/454 assembled read data: This feature allows to automatically correct some of the errors in NGS data from IonTorrent or 454 data. This requires that the assembled read data exists (i.e., ACE or BAM files) and that the Procedure Details were defined as IonTorrent/454.
- Assign new allele types for local typings: Only applicable if a Task Template defines an Allele typing query. The allele types are automatically queried, and if possible a new allele type is assigned to unknown allele sequences.
- Batch import mode: If enabled, the scanning of genes and storing Samples is performed without any user interaction. By default only empty targets in existing Samples are filled in batch mode. By selecting Overwrite existing targets also already filled targets are overwritten.
- Read Data Options: The read alignment can be imported from ACE- and SAM/BAM-files.
- Import read data for targets with analysis errors/warnings only: The read alignment data of targets where the analysis failed (red icon) or has warnings (yellow icon) will be kept. Alignment data of other targets will be discarded if not configured by other settings.
- Import read data for all targets: The read alignment data of targets will be kept.
- Matching Thresholds: Either use the thresholds that are defined by the Task Template(s) or manually define thresholds that should be used for ref.-seq. scanning.
- BLAST Options: The parameters that are used for BLAST when performing the ref.-seq. scanning. Default parameters are word size 11, mismatch penalty -1, match reward 1, gap open costs 5 and gap extension costs 2.
- MOB-suite Options: the use of MOB-recon can be disabled in this tab. In this case, contigs shorther than 500.000 bases are assumed to be a closed plasmid (e.g. PacBio data) and are directly passed to MOB-typer when the Chromosome and Plasmids Overview Task Template is used.
Click OK to start the process.
Scanning Targets in WGS data
Now the ref.-seqs. from the Task Template are scanned in the WGS data using the integrated BLAST. If a unique hit exists that succeeds the threshold that were defined in the Task Template or overwritten in the Scanning Procedure Details, the target is found.
![]() Hint: For SAM/BAM files (they contain reference-mapped data) a special consensus caller is used. Reads with a mapping quality below a given threshold (default 10) are discarded when the SAM/BAM file is read. The threshold can be set in the Preferences.
Hint: For SAM/BAM files (they contain reference-mapped data) a special consensus caller is used. Reads with a mapping quality below a given threshold (default 10) are discarded when the SAM/BAM file is read. The threshold can be set in the Preferences.
Preview the Found Targets
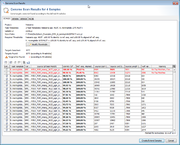
If the batch-mode was disabled, a table with all found hits is shown per input data file. Each row in this table represents one target that was searched. The rows that are highlighted red do not fulfill the defined thresholds.
Rows for targets that already exist in a Sample with the same name are disabled. To enable overwriting of existing target sequences, mark the checkbox Allow to replace existing targets.
The first column of the table shows a checkbox that defines if the found region should be extracted as sequence for the searched target. By default only the targets that fulfill thresholds unambiguously, and that are not already found in an existing Sample are selected.
The thresholds can be changed in this preview. The selection marks in the first column are updated automatically. The selection marks can also be changed manually row by row.
Press the confirm button at the bottom of the window to create the new Samples, or to extend existing ones.
Importing the Found Targets
Now the regions that match to the found targets are extracted from the input data, and added to new or existing Samples. If the input data contains the read information (ACE/BAM file), the aligned reads for this are also extracted and imported corresponding to the advanced settings. However, with default settings the read data will be discarded if the target succeeds all analysis checks to reduce the disk storage size.
FOR RESEARCH USE ONLY. NOT FOR USE IN CLINICAL DIAGNOSTIC PROCEDURES.