Contents
1 Overview
This tutorial describes how to use the Ridom Typer software to analyze a single target with Sanger sequence data without a reference sequence. If the target is very variable among various samples as it is the case for Internal Transcribed Spacer (ITS) sequences, it is recommended not to use a reference sequence.
2 Preliminaries
- This tutorial requires a running Ridom Typer client and server. Start the Ridom Typer server, then start the Ridom Typer client and initialize the database. For evaluation purpose a free evaluation license can be requested.
- Download and extract the example data archive Ridom_Typer_Examples_Sanger_ITS.zip
- The task template that is used in this tutorial requires Ridom Typer client version 10.0.2 or higher.
3 Downloading Task Template and Creating Project
- Step 1: The task template that is used here is not available via the Task Template store. Therefore, download the ITS UNITE task template manually as file.
- Step 2: Create a new Project for use with your sample data with the menu command File | New | Create Project
- Step 3: Enter a name in the field Project Name (e.g., ITS Tutorial). The fields Category and Acronym can be left empty.
- Step 3: Now press
 Add in Task Templates section to add a task template to the project.
Add in Task Templates section to add a task template to the project.
- Step 4: The dialog window Add Task Template to Project opens and shows the Task Templates that already exist in the database. Press the button File:Button16-Import.png Import from File and choose the downloaded file ITS_UNITE.tasktemplate
- Step 5: A editor window is shown containing with the ITS UNITE task template that will be imported. Further editings can be done here before importing. For the example data no further editing is required. Press OK to confirm the dialog and import the task template.
- Step 10: The newly imported task template is still selected in the task template dialog. Press OK to add the new Task Template to the Project. Then save your Project by confirming again with OK.
- Ridom Typer is a resequencing software. Once you have setup a project like this you can literally analyze hundreds/thousands of sequence data.
4 Assembling and Analysing Sanger Sequence Data
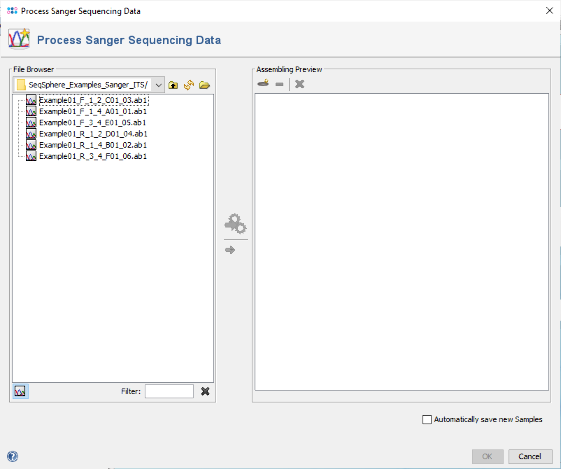
- Step 1: Choose from the menu File | Process Sanger Sequencing Data. Press the
 button above the file browser panel on the left, and choose the directory where you extracted the tutorial example data.
button above the file browser panel on the left, and choose the directory where you extracted the tutorial example data.
- Step 2: Select the tutorial example data directory or all six files in it, and press the button
 in the toolbar on the top of the right panel.
in the toolbar on the top of the right panel.
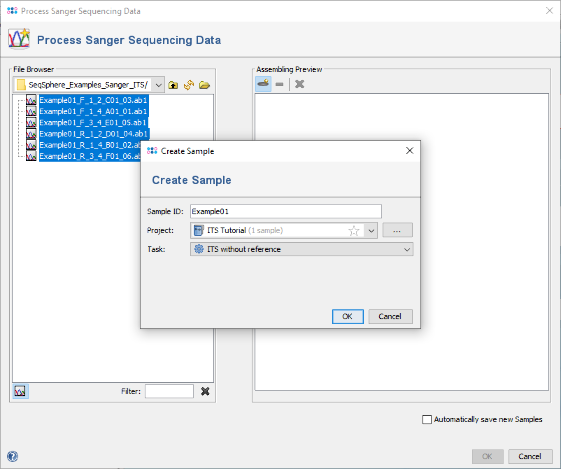
- Step 3: By default the Sample ID is extracted automatically from the shared parts at the beginning of the file names. (For more advanced file namings please see the MLST Sanger tutorial). Check if the correct Project is selected, and press OK.
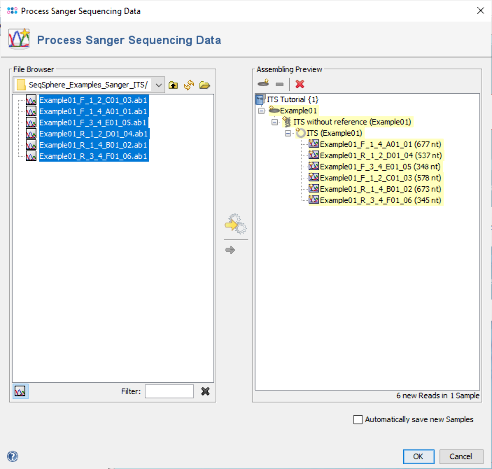
- Step 4: The preview on the right now shows the six chromatograms added to one newly defined Sample. Confirm the dialog to start assembling the chromatograms and processing the Sample.
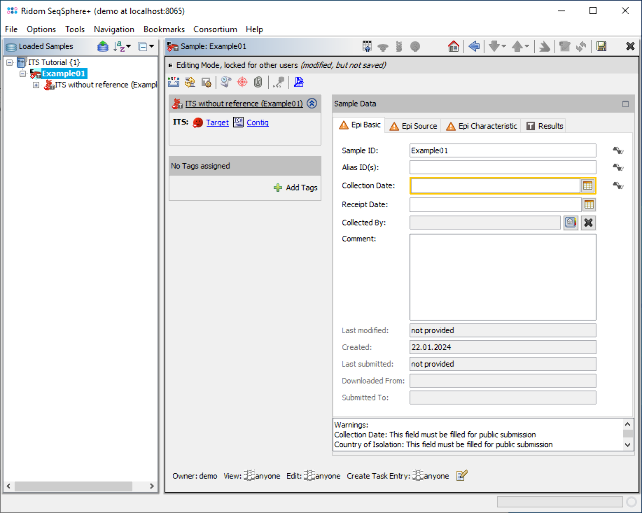
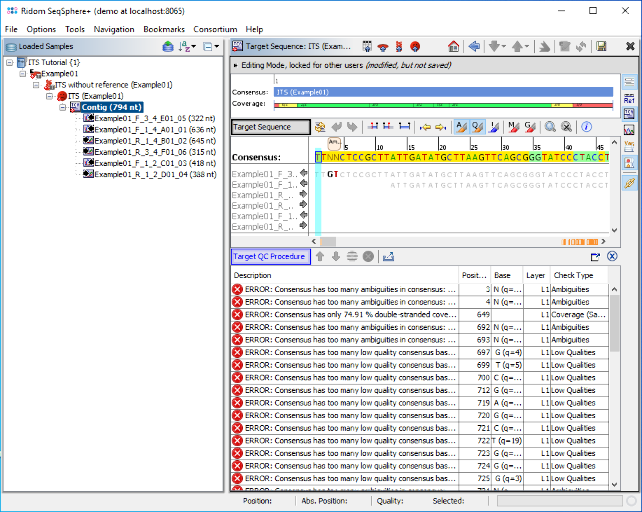
- Step 5: The six reads are now assembled into a new Sample listed in the tree on the left. On the right, an overview and the epi metadata are shown. The overview shows one ITS target. Click on the link Contig to show the assembled sequence data.
- Step 6: The contig alignment and the results of the Target QC Procedure are shown. The vertical toolbar on the right allows to show and hide different views for the sequence data. Press the button
 to show the chromatograms. Click on a position on the contig or use the button
to show the chromatograms. Click on a position on the contig or use the button  to synchronize the positions between the views.
to synchronize the positions between the views.
- Step 8: Double-click on the Task Entry node ITS UNITE (ExampleFungi) in the navigation tree on the left. The Task Entry overview is shown, with an empty Genotyping Results section on the right. The sequence was successfully determined as
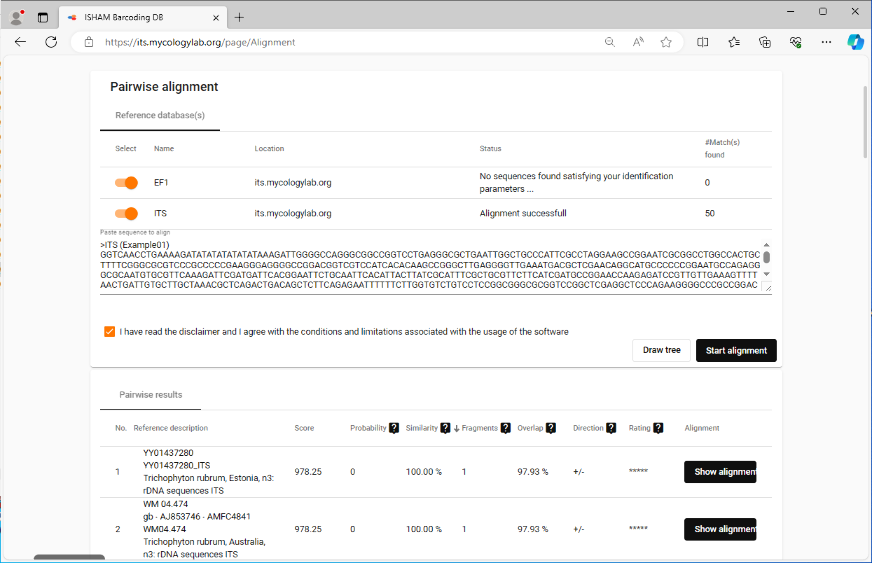
- Step 9: To use further web services, the consensus can be exported to clipboard or file by right-clicking on the consensus in the 'Target Sequence' section and choosing Copy Bases of Consensus Area(s) to Clipboard or Export Bases of Consensus Area(s) to File. For this tutorial, copy the consensus to the clipboard then go for example to the ISHAM Barcoding Database web service. Now paste in the copied consensus sequence and confirm with the Start alignment button to start the query. The closest hits will be shown on the website.
5 Storing and Retrieving Samples
- Step 1: Choose from the menu
 File | Save All to store the Sample to the database on your Ridom Typer server.
File | Save All to store the Sample to the database on your Ridom Typer server.
- Step 2: Choose File | Close All to remove it from the workspace
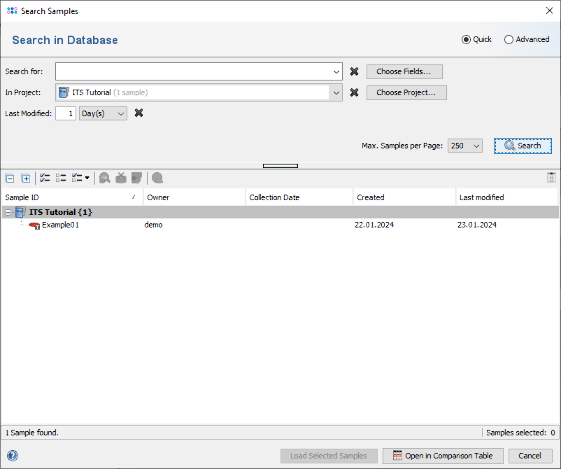
- Step 3: Choose
 File | Search Samples. Select the project in the Project box, and choose 1 day(s) for Recently modified. Then press the Search button.
File | Search Samples. Select the project in the Project box, and choose 1 day(s) for Recently modified. Then press the Search button.
- Step 4: The sample that just was saved is listed.