Contents
Introduction
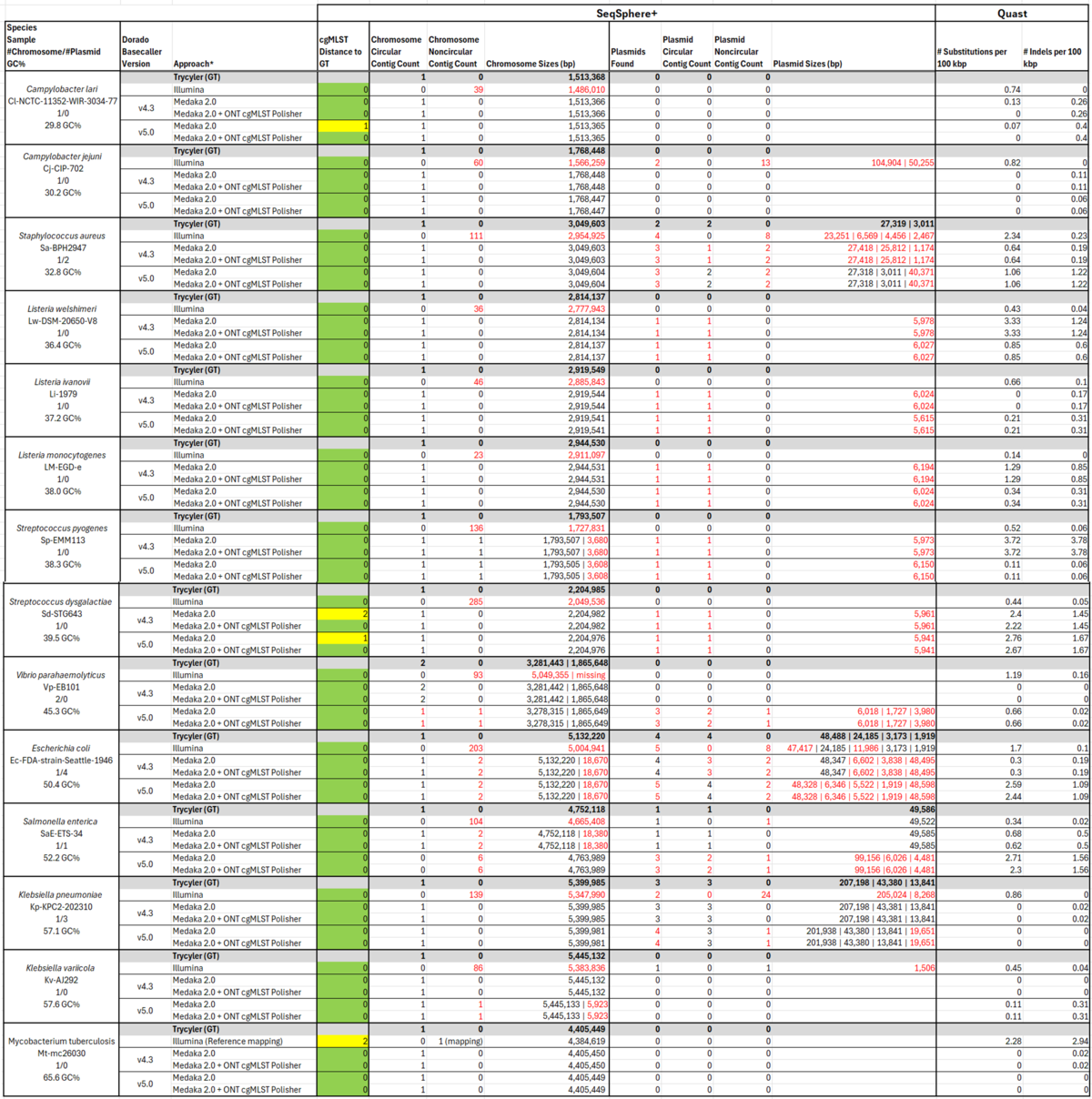
As we have shown in our accuracy evaluation, Medaka v2.0 and ONT-cgMLST-Polisher polishing improve the accuracy of ONT sequences substantially. Here, we additionally analyzed the contiguity of 14 bacterial genomes after Medaka v2.0 polishing.
Methods
We downloaded Illumina Fastq and ONT pod5 files of 14 bacterial genomes (GC range: 29.8 – 65.6%) published by Hall et al. (PMID: 39388235).
Illumina reads (2x 150bp, NextSeq2000 or MiSeq) were adapter trimmed, downsampled to 100x coverage and assembled with SKESA v2.3.0. The downloaded ONT (NBK/RBK) sequences were basecalled with Dorado v0.7.1 SUP 4.3 and 5.0 models (m5.0 was called from the downloaded pod5 files). Subsequently, the reads were trimmed for a quality of 10 and a minimum length of 500 with Chopper v0.7.0, downsampled to 100x coverage with Rasusa v0.8.0 --deterministic , and de novo assembled with Flye v 2.9.3-b1797 --nano-hq --deterministic. The assemblies of both m4.3 and m5.0 basecalling were polished with Medaka v2.0
(bacterial methylation model) with and without subsequent polishing using the ONT-cgMLST-Polisher. Ground truth reference assemblies were constructed using Trycycler v0.5.4 with ONT NBK/RBK SUP m4.3 and Illumina hybrid assemblies.
Contig counts and sizes were inferred from the Chromosome & Plasmid Overview. Substitutions and Indels were analyzed with Quast v5.3 using the Trycycler assemblies as reference.
Results
After polishing with the ONT-cgMLST-Polisher , the 14 isolates did not contain any errors on cgMLST level. A few errors on whole genome level were detected using Quast, showing differences between m4.3 and m5.0 basecalling. In addition to Medaka v2.0, we also analyzed Flye-only and Medaka v1.12 polished assemblies (data not shown) and didn't see any differences in terms of contig counts between the different polishing approaches. In the ONT assemblies, we found several samples with additional contigs but none with missing. In 4 cases, the contig count differed between m4.3 and m5.0 basecalled assemblies.
Table: cgMLST distance to the ground-truth, contig counts, chromosome and plasmid sizes, and number of substitutions and indels for 14 bacterial isolates. Deviations in contig counts as well as size differences of >1% in comparison to the Trycycler ground-truth are highlighted in red. Medaka 2.0 was run with the r1041_e82_400bps_bacterial_methylation model.
Conclusion
Although polishing with Medaka v2.0 and ONT-cgMLST-Polisher improves the accuracy of ONT sequences substantially, Flye assemblies are not always perfect and can lead to erroneous plasmids. Thus, if genome contiguity is of high importance, hybrid assemblies, e.g. constructed by Hybracter, are needed.
FOR RESEARCH USE ONLY. NOT FOR USE IN CLINICAL DIAGNOSTIC PROCEDURES.