Contents
Introduction
Here we compare the short-read data de novo assemblers that are supported by Ridom Typer. A specific focus is given to the end of the year 2018 published SKESA assembler. The authors from NCBI of the SKESA assembler claim in their publication (citation) that the assembler
- produces assemblies of equal or better quality than SPAdes,
- is at least 4x faster than SPAdes,
- produces identical results regardless of the number of threads or memory,
- scales well with increase in compute resources (minimum 16 GB RAM recommended) or coverage, and
- handles low-level contamination in reads (contiguity decrease when contamination level increases to 9x or above).
In this evaluation we aim to confirm their claims with a specific focus on allele calling efficiency.
Methods
Three strains with finished genomes were re-sequenced on an Illumina MiSeq machine. The strains cover the whole range of G/C content, i.e., low Staphylococcus aureus strain COL (NC_002951; 2.8 MBases genome), medium Escherichia coli strain Sakai (NC_009089; 5.5 MBases genome), and high Pseudomonas aeruginosa strain PAO1 (NC_002516; 6.3 MBases genome). 250bp Nextera XT paired-end (PE) libraries were produced for all 3 strains. In addition were for S. aureus 150bp and 300bp PE libraries constructed. Furthermore, the effects of BWA remapping for SKESA and SPAdes was evaluate with C. difficile strain DSM 27543 (NC_009089; 4.3 MBases genome using a cgMLST seed-only scheme with 2,270 targets). Finally, two 250bp PE libraries of mixtures with different concentrations of S. aureus and Enterococcus faecium strain ATCC BAA-472 (NC_017960.1; 3 MBases genome) were produced.
Before assembling the data were downsampled to different estimated coverages. Assembly of the produced data was performed with the three different de novo assemblers that are available in Ridom Typer: SKESA (version 2.3), SPAdes (version 3.11) and Velvet (version 1.1). For SKESA and SPAdes Ridom Typer performed a polishing step by remapping the reads with BWA and calling a new consensus on them. Velvet supports such a polishing step natively. For the resulting contigs an allele calling was done with Ridom Typer using cgMLST seed-only schemes based on the NCBI GenBank entry of the same strain. SKESA and SPAdes were run on Linux whereas Velvet was run on Windows.
The following parameters were compared for different coverages and assemblers:
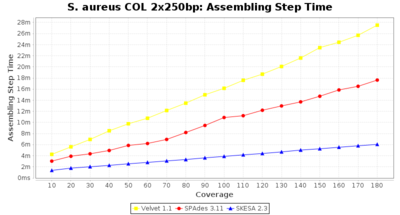
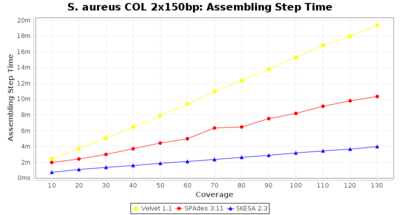
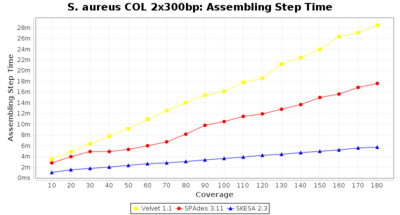
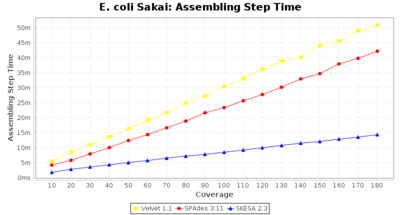
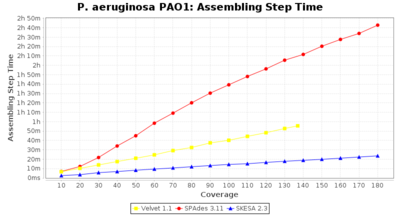
- Assembling Step Time
- The time the assembling step took (including BWA remapping) within the pipeline (smaller values are better) on an Intel Xeon system with 20 cores (two E5-2630) and 192 GB memory. The allele calling time is not counted here as it is anyway the same for all assemblers.
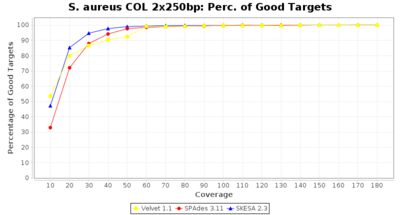
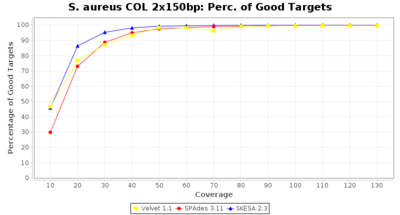
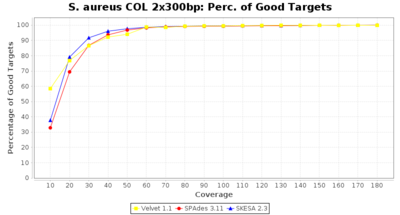
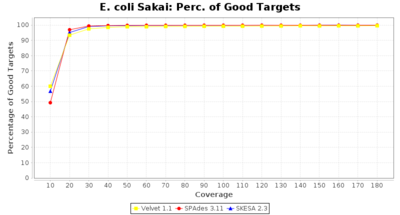
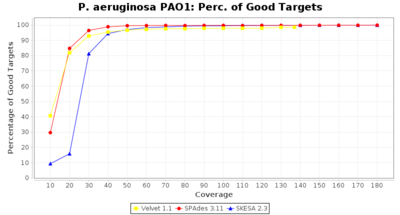
- Percentage of Good cgMLST Targets
- The number of targets that pass the quality check in a reference-only cgMLST task template that was defined with the downloaded NCBI genome of this strain (larger values are better).
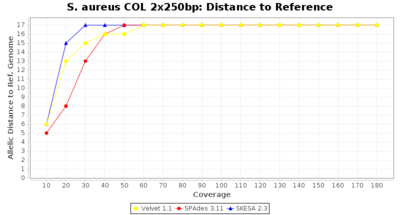
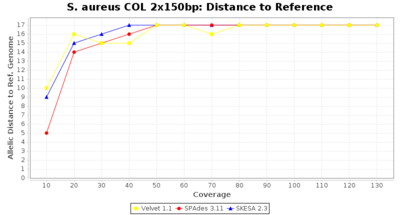
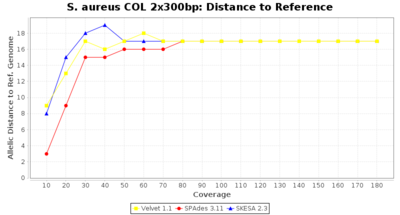
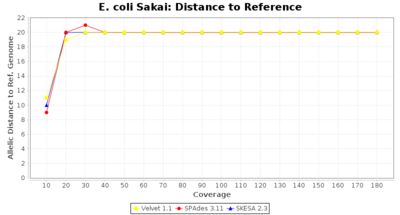
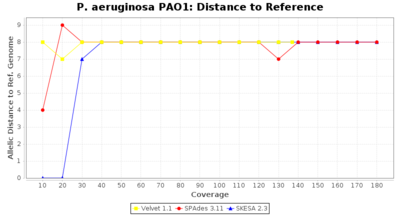
- Distance to Reference (Seed)
- The absolute number of differing alleles between the assembly and the downloaded NCBI genome of this strain (smaller values are better, but will never be zero due to sequencing errors in the finished NCBI genome and/or micro-evolutionary changes of the here sequenced culture collection strain during multiple sub-cultivation passages).
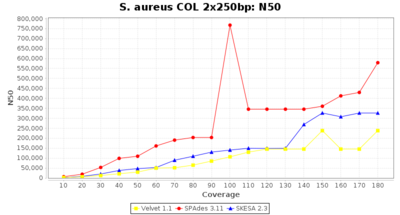
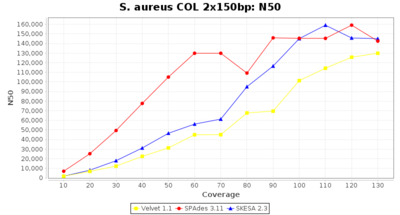
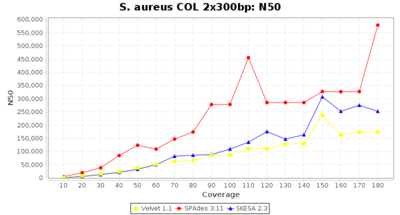
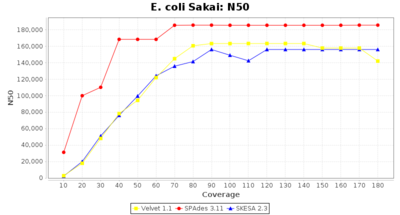
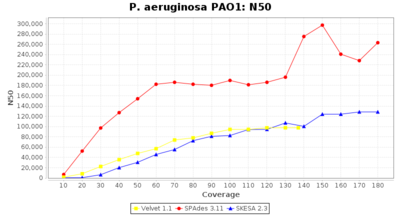
- N50
- The N50 value is a measure for assembly quality in terms of contiguity. N50 is described as a weighted median statistic such that 50% of the entire assembly is contained in contigs or scaffolds equal to or larger than this value.
Results
Assembler Time / Allele Calling Efficiency / N50 from Pure Culture
Staphylococcus aureus Ridom Typer used a cgMLST seed-only scheme with 2,486 targets.
Staphylococcus aureus 250bp PE
Staphylococcus aureus 150bp PE
Staphylococcus aureus 300bp PE
Escherichia coli 250bp PE
Ridom Typer used a cgMLST seed-only scheme with 4,225 targets.
Pseudomonas aeruginosa 250bp PE
Ridom Typer used a cgMLST seed-only scheme with 5,267 targets.
Effects of BWA Remapping
By default a remapping is performed in a pipeline that uses SPAdes or SKESA with the default settings. After the SPAdes or SKESA has finished, BWA is used to align the reads against the resulting assembly contigs. BWA mem is used as algorithm and for the resulting alignment (BAM file) a new consensus is called. As SPAdes is known to create inaccurate alignments at lower coverages, the consensus calling procedure is configured to use a threshold of 5 as minimum coverage to call a non-ambiguous base. For SKESA no minimum coverage is required by default. For both, the required read support for a consensus base is 60%.
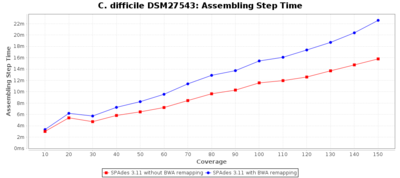
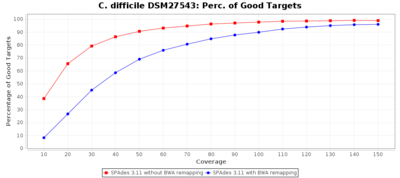
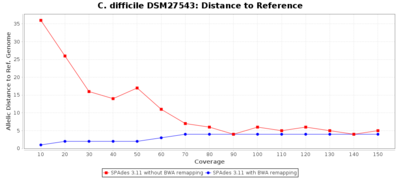
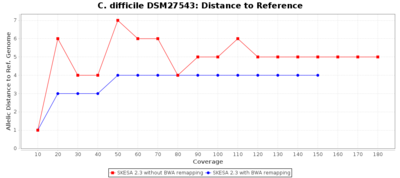
The following three charts show the effects for SPAdes on C. difficile with different coverages. With BWA remapping the percentage of good targets decreases somewhat, but most of the incorrect allele calls are corrected.
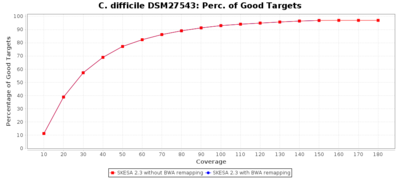
For SKESA the improvement by the BWA remapping is much smaller as shown in the following charts (note the difference in scale of the Distance to Reference chart).
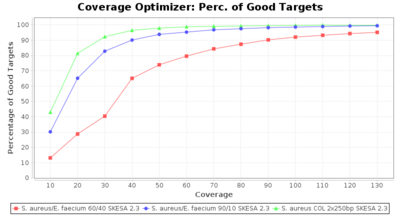
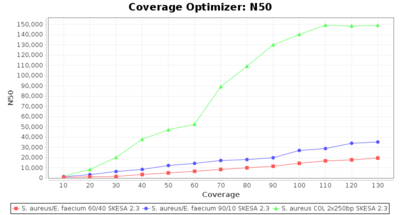
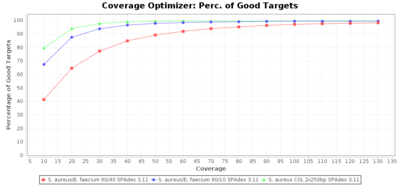
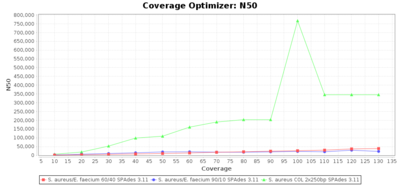
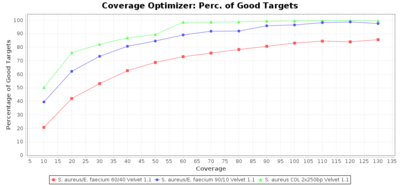
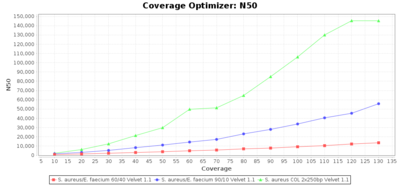
Percentage of Good cgMLST Targets / N50 from Mixed Culture
DNA of S. aureus strain COL (NC_002951; 2.8 MBases genome) and Enterococcus faecium strain ATCC BAA-472 (NC_017960.1; 3 MBases genome) were mixed with 60:40 and 90:10 ratios, respectively. Re-sequencing of 250bp PE Nextera XT libraries was done on a MiSeq. Resulting reads were downsampled to different estimated coverages relative to the COL genome size and processed with Ridom Typer using a Staphylococcus aureus cgMLST reference-only scheme with 2,486 targets. The SKESA and SPAdes assembler were run without BWA remapping. For comparison also the pure culture data of the COL strain are shown in the graphs. Indeed SKESA tolerates contaminations of up to 10% very well. Furthermore, are the contigs from the contaminant removed from the consensus assembly FASTA (data not shown).
SKESA
SPAdes
Velvet
Summary
The claims of the authors of the SKESA publication that were checked could be verified. SKESA is a very conservative assembler that tolerates up to 10% contaminating reads. Although not documented here, SKESA indeed scales very well with increasing compute power. In summary, SKESA produces very fast high quality de novo assemblies that are very well suited for cgMLST allele calling.
FOR RESEARCH USE ONLY. NOT FOR USE IN CLINICAL DIAGNOSTIC PROCEDURES.