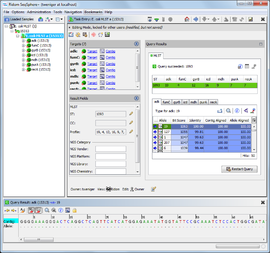
The Task Entry overview shows a summary of the genotyping results and the data that are stored within the Task Entry.
The view is organized in two columns:
- The left column shows a summary of all targets of this Task Entry, followed by the result fields. Usually the result fields are automatically filled by the genotyping query. The button
 Enable manual editing allows to manually fill or overwrite the values.
Enable manual editing allows to manually fill or overwrite the values.
- The right column shows the result details of the genotyping queries defined for this Task Entry. The layout of the result details are depending on the type of task template (e.g., cgMLST/Accessory Task Templates, Resistance/Virulence Task Templates, MLST Task Templates, Geno-Serotyping Task Templates). The column can be maximized/unmaximized by clicking the
 /
/ icon in the upper right corner (maximized can be the default for some task template, e.g. VFDB).
icon in the upper right corner (maximized can be the default for some task template, e.g. VFDB).
The toolbar on the top of the panel can shows the following buttons:
-
 Export Target Sequences: Export the consensus sequences of this Task Entries into a FASTA library file.
Export Target Sequences: Export the consensus sequences of this Task Entries into a FASTA library file.
-
 Reassemble Reads: Re-Assemble the read sequences (e.g., chromatograms) of all targets of this Task Entry.
Reassemble Reads: Re-Assemble the read sequences (e.g., chromatograms) of all targets of this Task Entry.  This feature is only available for Sanger sequencing data.
This feature is only available for Sanger sequencing data.
-
 Restart Genotyping: Restart the queries of this Task Entry.
Restart Genotyping: Restart the queries of this Task Entry.
-
 Position Navigator: Open the Position Navigator for this Task Entry.
Position Navigator: Open the Position Navigator for this Task Entry.
-
 Add Sequences from Assembly File: Open the Process Assembled Genome Data window with the WGS data file linked or attached to the Sample. The Task for the Task Entry is already pre-selected. This feature is only available for whole genome sequencing data.
Add Sequences from Assembly File: Open the Process Assembled Genome Data window with the WGS data file linked or attached to the Sample. The Task for the Task Entry is already pre-selected. This feature is only available for whole genome sequencing data.
-
 Add reads: Add reads from files manually.
Add reads: Add reads from files manually.  This feature is only available for Sanger sequencing data.
This feature is only available for Sanger sequencing data.
-
 Browse Website: Browse a website that is associated with this task template
Browse Website: Browse a website that is associated with this task template
-
 : Browse Publication: Browse a publication that associated with this task template.
: Browse Publication: Browse a publication that associated with this task template.
On the bottom, specific access rights can be set for the Task Entry.
For some task templates (e.g. MLST), an extra panel below the Task Entry overview shows the alignment between the contig and query library sequences that are selected in the query result table.
FOR RESEARCH USE ONLY. NOT FOR USE IN CLINICAL DIAGNOSTIC PROCEDURES.