Please Note: Working with Plasmid Databases requires the Long Read Module (LRM)
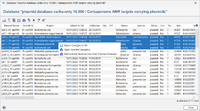
This windows lists all plasmids in a plasmid database for a Plasmid typing. The plasmid database is stored as a msh-file as an attachment to a Task Template.
The first column contains the plasmid name, the second column the Sample ID. The next columns contain the Project and the default Project metadata. Further columns contain the MOB-Suite results for a Sample.
The following functions can be used:
-
 Export the table.
Export the table.
-
 Create a snapshot of the database table. The snapshots can be opened using the menu command Tools > Comparison Table. Note: deleting database entries is not possible from a snapshot.
Create a snapshot of the database table. The snapshots can be opened using the menu command Tools > Comparison Table. Note: deleting database entries is not possible from a snapshot.
-
 Export plasmid contigs to file.
Export plasmid contigs to file.
-
 Open selected samples.
Open selected samples.
-
 Add additional database fields as columns.
Add additional database fields as columns.
-
 Visualize selected plasmids with pyGenomeViz. Metadata from the "Chromosome & Plasmid Overview" (rep(s) in green), "NCBI AMRFinderPlus" (AMR priority genes: red, other orange) and "CGE MobileElementFinder" (light blue) are included as genes in the visualization.
Visualize selected plasmids with pyGenomeViz. Metadata from the "Chromosome & Plasmid Overview" (rep(s) in green), "NCBI AMRFinderPlus" (AMR priority genes: red, other orange) and "CGE MobileElementFinder" (light blue) are included as genes in the visualization.
-
 Do a Single Linkage Clustering of selected plasmids.
Do a Single Linkage Clustering of selected plasmids.
-
 Show a Distance Matrix with the Mash distance for the selected plasmids.
Show a Distance Matrix with the Mash distance for the selected plasmids.
Note that it might take some moments to load the Sample data and MOB-Suite results for the Samples. Some functions are only available after this data was fetched.
Plasmids are not automatically removed from a plasmid database if a sample or a task entry containing the plasmid is deleted from the Ridom Typer server database. Therefore, there can be entries in the plasmid database without a Sample ID.
Selected entries can be permanently removed from the Mash database using the context menu command ![]() Permanently Remove Row from Plasmid Database. Note: In contrast to a similar named function in the plasmid table, the entries are permanently deleted from the database.
Permanently Remove Row from Plasmid Database. Note: In contrast to a similar named function in the plasmid table, the entries are permanently deleted from the database.
FOR RESEARCH USE ONLY. NOT FOR USE IN CLINICAL DIAGNOSTIC PROCEDURES.