The genomic allelic profiles form the basis of a portable, expandable, scalable, stable, and computer-readable nomenclature. Core genome MLST complex types (CT) function as an additional unique identifier for human communication purposes. Assigning to every combination of genomic alleles a unique identifier - similar like it is done for MLST sequence types (ST) – is not really helpful as nearly every sample exhibits a unique allelic profile. Therefore, similar to MLST clonal complexes (CC) CTs ‘lump’ together samples with very similar cgMLST profiles. Of course CT bins are much smaller than CC ones. CC founders are always defined ‘ad hoc’ during analysis as the ST with the most single locus variant (SLV) offspring STs. Thereby, the CC founder can change and clonal complexes can merge when the analysis is repeated at a later time point. In contrast, the CT founder status is assigned only once and is static to warrant for a stable nomenclature. Also in contrast to MLST CCs, the CT threshold is species-specific defined in the Task Template. Like for MLST STs or CCs the actual numeric CT value does not express relationship. The information content of CTs consists exclusively of being identical or non-identical.
The cgMLST.org nomenclature server assigns CTs during the sample submission process. If a submitted sample is less or equal than the CT threshold alleles distant to an already established CT founder it gets the CT value of this founder assigned. Otherwise a new CT is established and the sample becomes the founder of this CT. If a submitted sample is within the boundaries of two or even more CT founders, the sample gets the CT with the lowest numeric value assigned that not necessarily belongs to the closest CT founder. This tie-breaking rule was introduced in order to prevent the possibility of different CT assignments for identical allelic profiles depending on the order of submissions and time point of establishment of CT founders. Thus, thereby it is ensured that an exact identical allelic profile that is submitted multiple times always gets the same CT assigned. CT founder samples are currently not specifically marked. However, when searching the cgMLST.org server for a certain CT the sample listed with lowest cgMLST.org ID is the founder for this CT.
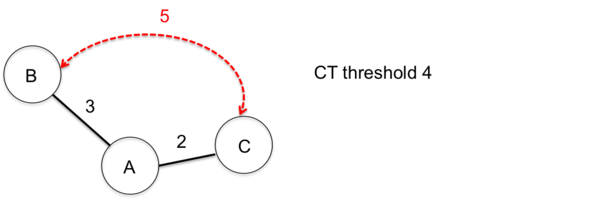
As the nomenclature is incremental expanding by definition and due to the entry order of samples and/or missing genotyping data, occasionally weird CT results can occur. In the following two such examples are illustrated for a CT threshold of 4.
Issue with entry order of submission
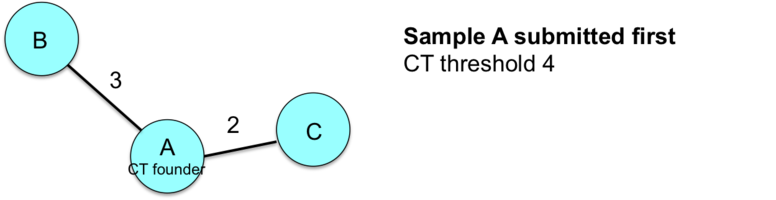
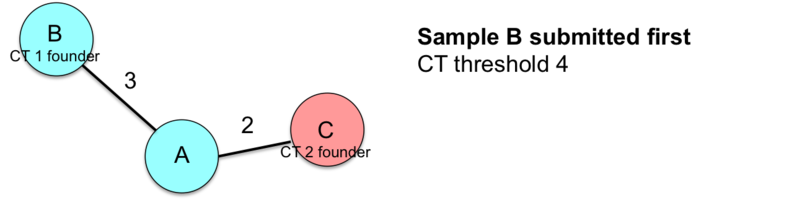
This example illustrates how the entry order of submissions influences the sample CT assignments.
![]() Important: Therefore, the index case and/or earliest isolate should always be submitted first to the nomenclature server if conducting a retrospective study.
Important: Therefore, the index case and/or earliest isolate should always be submitted first to the nomenclature server if conducting a retrospective study.
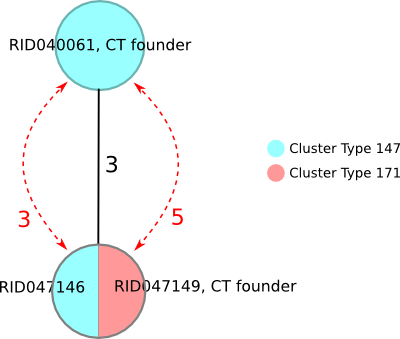
Issue with entry order that is complicated by missing genotyping data – two different CT fall together
This example illustrates how missing genotyping data may complicate the entry order issue. Sample RID040061 was submitted before the CT 171 founder and became founder of CT 147. By applying the tie-breaking rule of the lower CT numeric value, sample RID047146 got CT 147 assigned although falling together with the CT 171 founder. Only if the two ‘not found’ targets would be present and identical to the CT 171 founder when doing re-sequencing and -submission the CT would change to CT 171!
| Sample ID | Complex Type | lpg1099 | lpg1209 | lpg1618 | lpg2022 | lpg2506 |
|---|---|---|---|---|---|---|
| RID040061 | 147 | 13 | 19 | 23 | 2 | 29 |
| RID047146 | 147 | not found | 2 | not found | 30 | 2 |
| RID047149 | 171 | 2 | 2 | 2 | 30 | 2 |
The five differing targets shown in a comparison table.
Stability and reproducibility (even if reproducible done wrong) are of utmost importance for an incremental expanding nomenclature. However, as illustrated above cgMLST CTs are by no means perfect.