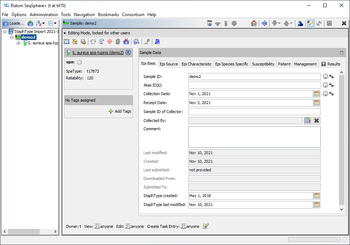
This FAQ describes how to import the database content of the by us no longer supported Ridom StaphType tool. The spa-types and epi metadata of such a database can be imported into Ridom Typer (requires client version 8.3 or newer and server version 8.3 or newer). When importing the StaphType database into Ridom Typer a new project named by default StaphType Import yyyy-mm-dd and a new database scheme will be created, that includes nearly all fields that were used in StaphType. The spa-types and the reliability rate are imported into result fields of a spa-typing task template. Therefore, the imported samples can be used in a comparison table and the distance for an MST will be calculated by the BURP distance of the spa-types. This import does not include chromatogram files or FASTA sequences.
Contents
Export from StaphType
![]() Important: The 'tx' spa-types, that were given by StaphType for local not submitted spa-types are not supported in Ridom Typer and will be handled as missing data in distance calculation! Therefore, be sure to submit all reliable tx-types with StaphType before exporting the database.
Important: The 'tx' spa-types, that were given by StaphType for local not submitted spa-types are not supported in Ridom Typer and will be handled as missing data in distance calculation! Therefore, be sure to submit all reliable tx-types with StaphType before exporting the database.
- Step 1: Start StaphType and login.
- Step 2: Invoke in the StaphType menu Database | Administration Tools | Create Special Seq.-Data Import/Export Filters and confirm the confirmation dialog.
- Step 3: Invoke in the StaphType menu
 File | Export Data to CSV File.
File | Export Data to CSV File.
- Step 4: In the "Export Entries to File" dialog, choose as Export Filter SeqData Export and press the edit button
 below the choosing box.
below the choosing box.
- Step 5: The "Edit Export Filter" window opens. In the "Selected fields" panel on the right scroll to the bottom and click on the entry SeqDataBLOB to select it. Then click the left arrow button
 to remove the entry from the list. Press OK to confirm the "Edit Export Filter" dialog.
to remove the entry from the list. Press OK to confirm the "Edit Export Filter" dialog.
- Step 6: Now choose the directory for exporting and press the button Save to export the fields of the database to the CSV file.
Import into Ridom Typer
- Step 1: Start Ridom Typer and login.
- Step 2: If not done yet, the spa-typing task template must be downloaded into the Ridom Typer database: Choose in the Ridom Typer menu Options | Task Templates. Then press the button Download to open the Task Template Sphere dialog. Choose Staphylococcus aureus, select the spa-typing task template and confirm the download. The task templates dialog can be closed then.
- Step 3: Invoke in the Ridom Typer menu File | Open File(s) and choose the CSV file that was exported above.
- Step 4: In the Open Files dialog click on the item Import StaphType Database and change or confirm the default name of the new project that will be created for the StaphType database. Confirm with the OK button to start the import. The import is done in the background and takes about 3 seconds per sample.
Future Project Management
A user may continue with new S. aureus samples (e.g., from WGS data) stored in the just created project. Alternatively, a new project using the just created database scheme (named also by default StaphType Import yyyy-mm-dd) or the default bacteria database scheme may be used. Samples from various projects can be analyzed together in one comparison table.
The search for similar samples can be used to find for selected samples other samples in the database, that have the same or a similar spa-type. Two spa-types are regarded as similar if the BURP distance is equal or below 4 and only spa-types with at least five repeats can be compared for similarity.
An early warning alert can be defined for the project, that triggers an alarm if samples with the same spa-type as existing ones are newly processed. Several metadata criteria can be set for the alert.
Details about Field Mapping
As far as appropriate the fields of the Ridom Typer database scheme Staphylococcus aureus are used to import the data of the StaphType fields, i.e.:
| From StaphType Field(s) | Into Ridom Typer Default Field(s) |
|---|---|
| Isolate ID | Sample ID |
| Order ID | Alias IDs |
| Isolation Date | Collection Date |
| Receipt Date | Receipt Date |
| Internal Comment and Sync Comment | Comment |
| Origin | Source Type and Source Subtype |
| Specimen | Isolation Source |
| Care Unit ZIP, City State, Country | ZIP, City State, Country of Isolation |
| Case ID | Case ID |
| PubMed ID | PubMed ID(s) |
| MRSA/MSSA | MRSA/MSSA |
| Report Comment | Report Comment |
For other fields that were filled in StaphType for at least one sample the import creates new fields automatically: Additional Strain Source fields are imported into new fields under the tab Epi Source. Additional Strain Type fields are imported into new fields under the tabs Epi Characteristics and Epi Species Specific. The user-defined fields of Strain Source and Strain Type are imported into new fields under the tab Epi Characteristics. The Susceptibility and Management fields are imported into new fields under two tabs with the same names. Finally, the Case fields are imported into new fields under the tab Patient.
Not imported into Ridom Typer are the StaphType fields: SpaServer Submitter, Care Unit contact fields, Clinic and Ward address and contact fields (but Clinic/Ward names are imported!), Patient address fields, Isolation scheme, Person ID(s) of patient contacts, Intra Hopsital Transfers, and Report-receiver. Furthermore, as mentioned already above chromatogram files, other sequence data, and reports are not imported into Ridom Typer.
![]() Hint: It is recommended to keep for potential future use the above exported CSV file and a backup of the StaphType database at a safe place. To create a database backup, invoke in the StaphType menu File | Make Database Backup and choose the directory where the backup file should be stored.
Hint: It is recommended to keep for potential future use the above exported CSV file and a backup of the StaphType database at a safe place. To create a database backup, invoke in the StaphType menu File | Make Database Backup and choose the directory where the backup file should be stored.