| wiki | search |
Uploads the MIRU-VNTR data to the MIRU-VNTRplus website and does lineage identification using best matching strains there. Identification is done using categorical distance for all 24 MIRU-VNTR loci.
The reference database checks all best matching reference strains that have a distance lower than the given match distance. If the lineages of all these best matching strains are the same, they are copied to the table, together with the calculated match distance.
To make sure that the MIRU-VNTR columns have he correct names, the command Rename MIRU-VNTR columns to default can be used.
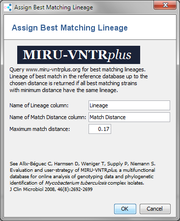
- Name of Lineage column: Enter a name for the column in which the retrieved lineage information is stored. An existing column can be used, but existing values will not be overwritten.
- Name of Match Distance column: Enter a name for the column in which the distance to the best matching reference database strains is stored. An existing column can be used, but existing values will not be overwritten.
- Maximum match distance: a maximum match distance of 0.17 is suggested. If this values does not identify enough lineages, a relaxed value of 0.3 is suggested. With the relaxed value more strains are identified, but there might be a few wrong identifications (Allix-Béguec et al. 2008).
References
- Allix-Béguec C, Harmsen D, Weniger T, Supply P, Niemann S. Evaluation and user-strategy of MIRU-VNTRplus, a multifunctional database for online analysis of genotyping data and phylogenetic identification of Mycobacterium tuberculosis complex isolates. J Clin Microbiol 2008, 46(8):2692-2699 Pubmed.