BWA is a reference mapper for short read data. It maps short reads to a given reference genomes. BWA contains several algorithms, the implementation in SeqSphere+ uses the bwa-sw algorithm (version 0.6.1/0.6.2 for windows/linux).
![]() Hint: Normally the reference mapper is used as part of the SeqSphere+ Assembling Pipeline.
Hint: Normally the reference mapper is used as part of the SeqSphere+ Assembling Pipeline.
The BWA Reference Mapper can be directly accessed using the menu Tools | Short Read Mapper (BWA-SW).
The reference mapper reads from FASTQ-files that contain either single or paired reads. The input files can be quality trimmed and downsampled before mapping. The mapped reads and the reference sequence are stored in a BAM-file.
Reference mapping will work with most short read data. Compared to de-novo assembling it is fast and needs only a limited amount of memory. However, results depend on the chosen reference sequence. Therefore, reference mapping produces good results for monomorphic organisms (e.g. M. tuberculosis) or for Samples that are closely related to the isolate from which the reference sequence is taken.
Contents
Quality Trimming
The reads can be processed before they are mapped. They can be automatically trimmed based on read quality and downsampled.
BWA does its own trimming, so quality trimming is disabled by default.
When trimming is enabled, reads are trimmed on both ends until the average base quality is better than the given value in a window of a selected number of bases.
Downsampling
To reduce the size of the output files and the time required for mapping, the input files can be downsampled. Downsampling randomly removes reads so that the given approx. size is obtained. If quality trimming is selected, downsampling is done on the trimmed reads.
Depending on sequencing technology and read length different downsampling settings might be useful. For Illumina HiSeq/MiSeq data a downsampling to approx. 180 times the expected genome size worked well.
Standard Settings (in pipeline and manual mode)
- Quality trimming: reads can be trimmed from both ends until their quality is above the given average quality within a given window.
- Downsampling: select which coverage of expected genome size should be reached by downsampling.
- Threads: define the maximum number threads that should be used by bwa.
- Executable: the path to the bwa executable.
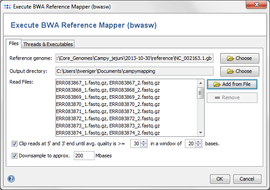
Manual Settings (in manual mode only)
- Reference genome: the genome that is the base for mapping. A FASTA or a GENBANK-file can be used as reference.
- Output directory: the directory where the resulting BAM-files will be written to.
- Read Files: the read files, in FASTQ or FASTQ.GZ format. Multiple files can be selected, and they can be automatically grouped by their filename (e.g. forward and reverse reads can be grouped together).
- Files contain paired reads: Check if the files contain paired reads. The mapper uses paired reads if exactly two files are in each file group. Both forward and reverse files must contain the same amount of reads, and read number X in the forward file must correspond to read number X in the reverse file.
- Threads: define the maximum number threads that should be used by bwa.
- Additional Options: specify additional command line options for bwa (see bwasw section at http://bio-bwa.sourceforge.net/bwa.shtml#3).
Postprocessing
Before a BAM file can be scanned by SeqSphere+, a consensus sequence is automatically calculated by the built in consensus caller.
Speed and Memory
Reference mapping does not require huge amounts of memory, 4GB is usually sufficient. Depending on genome size, read count, and read lengths reference mapping may take from 10 to 40 minutes or longer on a fast quad-core computer (in 2013).
| Species | Genome size | Coverage | Read length | Runtime |
|---|---|---|---|---|
| M. tuberculosis | 4.4 MBases | 150x | 2x 150bp | 10min |
| M. tuberculosis | 4.4 MBases | 250x | 2x 250bp | 14min |
| M. tuberculosis | 4.4 MBases | 270x | 2x 51bp | 16min |
Note that actual time requirements may be different depending on genome size, read length, coverage, read distribution, and read quality.
Open Source note
The BWA reference mapping function is a wrapper to an external BWA executable. This BWA executable is open source software that is licensed under the GNU General Public License version 3.0 (GPLv3). The program homepage is: http://bio-bwa.sourceforge.net/. The windows version of BWA is from here: http://bow.codeplex.com/. The source code is installed next to the executable.
For more information on BWA, see: Li H. and Durbin R. (2010) Fast and accurate long-read alignment with Burrows-Wheeler Transform. Bioinformatics, Epub. [PMID: 20080505]