Contents
cgMLST.org Nomenclature Server
The cgMLST.org Nomenclature Server (www.cgMLST.org) provides a global nomenclature for stable public cgMLST schemes, i.e. for Task Templates that were downloaded from the Task Template Sphere. If Samples are using those downloaded Task Templates they can be submitted to cgMLST.org.
![]() Important: If no submission of any data is wanted or allowed, then only local Task Templates must be used. Samples that are using only local Task Templates will not and cannot be submitted to cgMLST.org. Local Task Templates can be defined using the cgMLST Target Definer, or by converting public Task Templates into local ones.
Important: If no submission of any data is wanted or allowed, then only local Task Templates must be used. Samples that are using only local Task Templates will not and cannot be submitted to cgMLST.org. Local Task Templates can be defined using the cgMLST Target Definer, or by converting public Task Templates into local ones.
Allele Submission to cgMLST.org
When Samples are processed for a downloaded Task Template, the new alleles are by default automatically submitted to and stored at cgMLST.org. The cgMLST.org Nomenclature Server assigns new global allele type numbers to the alleles. The allelic profile is not stored at cgMLST.org during allele submission.
The automatic allele submission can be disabled in the in the client settings, by using the menu item Options | Preferences, and selecting the the item Online Connection | Allele Submission to cgMLST.org. However, if a Sample has new alleles that are not known at cgMLST.org, the new alleles are treated as missing data in the distance calculation (e.g., in Comparison Table).
Sample Submission to cgMLST.org
The Sample submission to cgMLST.org is optional (can be turned off in the assembly pipeline). It can be used to submit and store the allelic profile and optional metadata on cgMLST.org. Various options exist:
- Store only new CT founders as anonymized samples on cgMLST.org (default)
- Only the allelic profile and no further data (including submitter data) are stored on cgMLST.org
- Store only new CT founders on cgMLST.org
- Samples that are new CT founders are stored non-anonymized on cgMLST.org
- Store all submitted samples on cgMLST.org
- All Sample are stored non-anonymized on cgMLST.org
One of the three options is required to assign to all samples a Complex Type (CT). Choosing one of these options requires a one-time registration to the cgMLST.org nomenclature server.
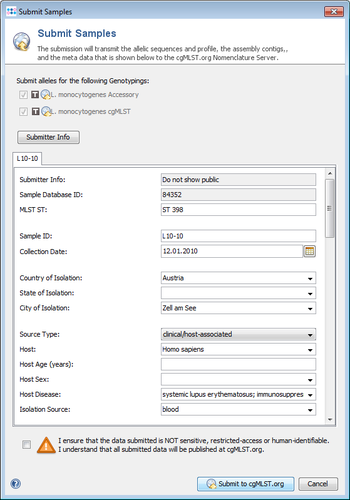
The submission of a non-anonymized Sample, i.e., option 2 or 3 above is selected, requires the transfer of the following minimum data:
- Sample ID (alternatively 'Alias ID' or 'do not submit and use cgMLST.org ID')
- Submitter Info (not shown public by default)
- Submission Date (only year is shown public)
- MLST Sequence Type (ST)
- Core and accessory genome MLST allelic profiles
- cgMLST Complex Type (CT, returned from cgMLST.org)
- Percentage of Good cgMLST Targets
- Genus
- Species
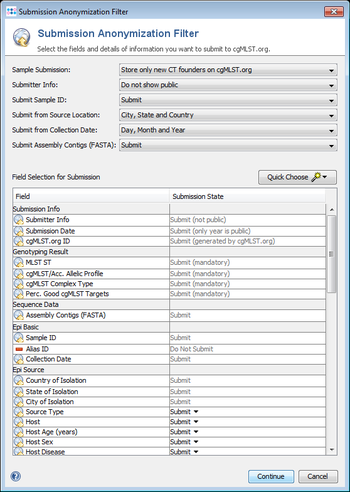
The amount of additional metadata that should be submitted can be defined in the Submission Anonymization Filter.
Submission to cgMLST.org is only possible if the Sample has in the cgMLST scheme at least 90% of good targets (i.e., targets passed the QC procedure and have therefore a green or yellow smiley). Allele types are only received for the good targets (green or yellow smiley). During the first submission, the user must register once with basic contact information. This information can later be modified using the menu function Options | User Settings. The submitter contact information is by default not public shown on cgMLST.org, but it will be used to forward incoming requests about submitted Samples.
Except for the submitter information and exact submission date any submitted data is immediately made publicly available on cgMLST.org. A unique serial number is assigned to the sample, and stored back as cgMLST.org ID in the local database. This number is also shown as link in the upper left corner of the Sample Overview. This cgMLST.org ID link can be used to directly access the public web page of the submitted Sample and to control the data that was submitted (e.g. RID002433).
With default settings, only new CT founders will be stored as anonymized samples on the cgMLST.org Nomenclature Server. The default settings can be changed in the general preferences settings.
The submission of Samples can be invoked manually or it can be configured in a pipeline script.
Manual Submission of a Small Number of Samples
If only a small number of samples should be submitted (e.g., around ten), it is recommended to use the following steps to see a full preview of the data that will be submitted.
- Step 1: Load the Sample(s) into the workspace using the menu function File | Search Samples.
- Step 2: Invoke the menu function
 File | Assign or Submit new Alleles and select all Samples that should be submitted.
File | Assign or Submit new Alleles and select all Samples that should be submitted.
- Step 3: The Submission Anonymization Filter dialog is shown to define which fields should be submitted. The last used setting is kept.
- Step 4: Finally the Submission Preview dialog is shown with the filtered data that will be submitted. The data can now be controlled and modified just before sending it. When the dialog is confirmed, the shown data is send to cgMLST.org.
Manual Submission of a Large Number of Samples
Use the menu function Tools | Manage Submissions to cgMLST.org or the submit button ![]() in the Search Samples dialog to submit multiple Samples at once.
The Submission Anonymization Filter dialog is shown to define which fields should be submitted. When the dialog is confirmed, the shown data is send to cgMLST.org. No preview of the submitted data will be shown when using this function.
in the Search Samples dialog to submit multiple Samples at once.
The Submission Anonymization Filter dialog is shown to define which fields should be submitted. When the dialog is confirmed, the shown data is send to cgMLST.org. No preview of the submitted data will be shown when using this function.
Automatic Submission via Pipeline
If Samples are imported through a pipeline, it can be configured in the Submission part of the Pipeline Script that the Samples are submitted to cgMLST.org. The Submission Anonymization Filter settings are defined for each pipeline script.
In contrast to the manual submission, the pipeline submission does not show a preview dialog before the samples are submitted, because all pipeline processing runs in an automatized way. Therefore, the button Preview Epi Data can be used to show the potentially submitted epi data of the matching samples that already exist in the SeqSphere+ server database.
Resubmission of Submitted Samples
If a Sample that was already submitted is submitted again, then the data at cgMLST.org is immediately updated. Fields that are filtered out or set to empty in the re-submission are immediately removed from cgMLST.org.
The procedure for re-submitting Samples is the same as the manual submission described above section.
Withdrawal of Submitted Samples
Submitted Samples can be withdrawn from the cgMLST.org Nomenclature Server.
Use the menu function Tools | Manage Submissions to cgMLST.org or the withdraw button ![]() in the Search Samples dialog to withdraw submitted Samples from cgMLST.org.
in the Search Samples dialog to withdraw submitted Samples from cgMLST.org.
Filtering Metadata for Submission
The level of metadata that should be submitted can be defined in the Submission Anonymization Filter that is shown in a dialog before a manual submission is confirmed. Also in a pipeline script the Submission Anonymization Filter settings can be configured if automatic submission is selected.
New epidemiological fields that are created by the user are never submitted.
Potentially the following metadata is submitted to cgMLST.org and shown on the public website. Unless the fields are mandatory, they can be excluded from submission (Do Not Submit). For place and time information additionally the level of detail can be set. The table shows the sample metadata fields (by default only of samples that are CT founders) that are submitted by default for a new installation of a SeqSphere+ Client.
| Submission Info | |
|---|---|
| Submitter Info | Submit (mandatory, not public shown by default) |
| Submission Date | Submit (mandatory, only year is public) |
| Sample cgMLST.org ID | Submit (generated by cgMLST.org) |
| Genotyping Result | |
| MLST ST | Submit (mandatory) |
| cgMLST/Acc. Allelic Profile | Submit (mandatory) |
| cgMLST Complex Type | Submit (mandatory) |
| Perc. Good cgMLST Targets | Submit (mandatory) |
| Sequence Data | |
| Assembly Contigs (FASTA) | Submit |
| Epi Basic | |
| Sample ID | Submit |
| Alias ID | Do Not Submit (alternative for Sample ID) |
| Collection Date | Submit |
| Epi Source | |
| Country of Isolation | Submit |
| State of Isolation | Submit |
| City of Isolation | Submit |
| Source Type | Submit |
| Host | Submit |
| Host Age (years) | Submit |
| Host Sex | Submit |
| Host Disease | Submit |
| Isolation Source | Submit |
| Epi Characteristic | |
| Genus | Submit (mandatory) |
| Species | Submit (mandatory) |
| Strain | Submit |
| Genotype | Submit |
| Serotype | Submit |
| Pathotype | Submit |
| Identification Method | Submit |
| Identification Kit Vendor | Submit |
| Culture Collection | Submit |
| PubMed ID(s) | Submit |
| Nuccleotide Accession(s) | Submit |
| Experiment Accession | Submit |
| Sample Accession | Submit |
| StudyAccession | Submit |
| Imported from EBI/NCBI1) | Submit (mandatory) |
| Epi Species Specific | |
| PFGE Pattern(s) | Submit |
| Epi Species Specific (only for MTBC) | |
| Spoligo | Submit |
| MIRU 15-9 Type | Submit |
| MIRU Lineage | Submit |
| Gagneux Lineage | Submit |
| Laboratory Procedure Details | |
| Nucleic Acid Extraction | Submit |
| Library Source | Submit |
| Library Strategy | Submit |
| Library Selection | Submit |
| Library Construction Method | Submit |
| Library Amplification Method | Submit |
| Sequencing Protocol | Submit |
| Library Insert Size | Submit |
| Sequencing Length | Submit |
| Sequencing Vendor | Submit |
| Sequencing Platform | Submit |
| Assembly Procedure Details | |
| Assembly Pre-processing | Submit |
| Assembly Type | Submit |
| Mapping Reference Genome | Submit |
| Assembler | Submit |
| Assembler Version | Submit |
| Assembler Parameters | Submit |
| Read Statistics | |
| Avg. Coverage (Unassembled) | Submit |
| Avg. Read Length (Unassembled) | Submit |
| Avg. Read Length (Processed, Unassembled) | Submit |
| Assembly Statistics | |
| Contig Count (Assembled) | Submit |
| N50 (Assembled) | Submit |
| Consensus Base Count (Assembled) | Submit |
| Avg. Coverage (Assembled) | Submit |
| Genome Status1) | Submit (mandatory) |
1) The fields Imported from EBI/NCBI and Genome Status are automatically filled if the data was imported from NCBI Genomes or SRA. Imported from EBI/NCBI can be true or false. The Genome Status can have one of the following values: Complete Genome, Chromosome, Scaffold, Contig, or SRA.