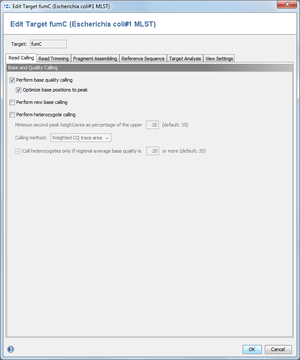
This dialog defines, if SeqSphere+ should perform quality/base/heterozygote callings on all chromatograms that are added to a target.
![]() This feature is only available for Sanger sequencing data.
This feature is only available for Sanger sequencing data.
Perform base quality calling
SeqSphere+ can calculate base qualities for each base call in the reads. The quality values are related to the error probability of the base calls. This operation will overwrite any existent base qualities that were calculated before and stored in the read files. It is recommended to enable this setting.
- Optimize base positions to peak
- Several base callers are not defining the base call position at the center of the peak, but to its upper left position. Because the Ridom base quality is calculated for the exact position, this may result in false quality values. Therefore, SeqSphere+ can correct the position of the base call to get a better quality guess for the peak. It is highly recommended to enable this option.
Perform new base calling
SeqSphere+ has a built-in algorithms to perform a base calling on chromatogram files. However, it is recommend to use the standard base-calling software (if available) that shipped with your sequencing machine, unless your results are poor.
Perform heterozygote calling
SeqSphere+ can detect heterozygote base calls in chromatograms. The heterozygote bases are called as ambiguities. The parameter for the second peak height/area can be used to configure the detection of the mixed base positions. By using the last option the heterozygote caller will be instructed to call only heterozygote bases, if a certain minimum regional (10 bases before and after the position) average base quality is surpassed.