Velvet is a de novo assembler for read data. Velvet works well for assembling reads from Illumina systems with read length from 70bp to 300bp length. However, Velvet does not work well with reads that contain many InDel-errors and is therefore not suited for assembling data from Ion Torrent or 454 machines.
Velvet (version 1.1.04, citation) is only available for the Microsoft Windows version of the Ridom SeqSphere+ Client.
![]() Hint: Normally the Velvet assembler is used as part of the SeqSphere+ Assembling Pipeline.
Hint: Normally the Velvet assembler is used as part of the SeqSphere+ Assembling Pipeline.
The assembler's algorithm is based on a so-called 'k-mer' data structure. The assembler results strongly depend on the chosen value for k. An automatic mode is included in SeqSphere+ that runs Velvet for multiple k-values and uses the results from the best run. Multiple Velvet processes can be run in parallel with different values of k to speed up the assembling. Velvet can require large amounts of memory, therefore at least 32GB of memory are recommended.
The Velvet assembler can be directly accessed using the menu Tools | Read Assembler (Velvet).
The assembler reads from FASTQ-files that contain either single or paired reads. The input files can be quality trimmed and downsampled before assembling. The assembled reads are written into an ACE-file, they can be imported using Process Assembled Genome Data.
Contents
Quality Trimming
The reads are processed before they are assembled. They can be automatically trimmed based on read quality and downsampled.
Default settings for trimming are to trim on both ends of the reads until the average base quality is > 30 in a window of 20 bases. This settings usually work well for Illumina HiSeq/MiSeq data. For other sequencing technologies different values might be used. Note that quality trimming will not remove a read with quality below threshold in all bases, but will always leave (2*window size) bases in the middle of the read.
It is recommended to enable quality trimming, it usually results in better assemblies.
Downsampling
To reduce the size of the output files and time and memory usage, the input files can be downsampled. Downsampling randomly removes reads so that the given approx. size is obtained. If quality trimming is selected, downsampling is done on the trimmed reads.
Depending on sequencing technology and read length different downsampling settings are useful. For Illumina data downsampling to approx. 120x coverage of the expected genome size works well. However, downsampling can be disabled or coverage modified if wished.
Choose of k-values
Velvet uses a 'k-mer' data structure for assembling. Assembling results differ based on the value for k. A good value for k can be vaguely guessed from the coverage and the read length. To get good assembling results, it is suggested to run Velvet using different values for k and return the assembly with the best results (measured by avg. length of contigs with more than 1000bp).
An automatic mode exists that uses an heuristic based on read length to determine k-values that should be checked. Velvet is then run for these k-values. The automatic mode usually finds good results, and using it is recommended for most users. Alternatively, an upper and a lower limit for k-values and a step size can be specified. Velvet is then started for all k-values in the given range and the best assembly is returned. If the computer used has enough RAM and cores available several k-mer values can be processed in parallel and thereby speeding-up the whole process.
Automatic mode heuristic
- Start on the lowest value of
- average read length/2
- 107
- End at the lowest value of
- average read length * 0.8
- 171
- Search first using k-mer step size of 8, then around the best hit with step size 2.
Postprocessing
The AMOS file that is created by Velvet is automatically converted to a ACE file format. The consensus bases are newly assigned by a consensus caller by SeqSphere+. The AMOS/ACE files do not contain base qualities for consensus or read bases.
Settings
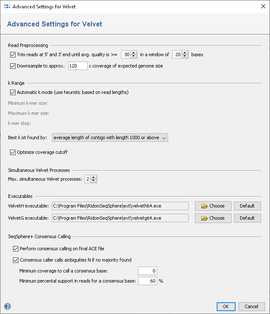
The dialog window allows to change settings for
- Output directory: the directory where the resulting ACE-files will be written to.
- Read Files: the read files, in FASTQ or FASTQ.GZ format. Multiple files can be selected, and they can be automatically grouped by their filename (e.g. forward and reverse reads can be grouped together).
- Paired Reads: Check if the files contain paired reads. The assembler uses paired reads if exactly two files are in each file group. Both forward and reverse files must contain the same amount of reads, and read number X in the forward file must correspond to read number X in the reverse file.
- Expected genome size: The expected size of the assembled genome (for downsampling).
- Quality trimming: reads can be trimmed from both ends until their quality is above the given average quality within a given window.
- Downsampling: select which coverage of expected genome size should be reached by downsampling.
- k Range: The minimal value for k, the maximum value and the step can be specified. Note that the k-values must be odd, and the step must be even. When automatic mode is used, k-values to check are calculated automatically.
- Optimize coverage cutoff: Do coverage cutoff optimization for each Velvet run. This usually improves the resulting output.
- Simultaneous Velvet Processes: The number of simultaneous processes can be specified. Note that running multiple Velvet processes in parallel will result in higher memory consumption. See paragraph Speed and Memory.
- Consensus caller calls ambiguities N if no majority found (checked by default): SeqSphere+ always performs an own consensus calling for Velvet assemblies that improves the results. If the sum of qualities of reads that contain the resulting base at the position is less than 60% of the total sum of read qualities at the position, an ambiguity 'N' is returned. If no quality values are given, the base count sums are used. If the checkbox is not checked the original Velvet consensus base is taken.
Speed and Memory
Memory usage of Velvet depends on
- number of reads
- read length
- read qualities
- read distribution (depends amongst others on used chemistry)
- genome size
- k-value
If possible, quality trimming and downsampling is recommended to reduce memory consumption. When running multiple processes of Velvet in parallel, memory consumption increases.
| Species | Genome size | Coverage | Read length | Total automatic mode runtime | Used memory for single Velvet process | Total used memory |
|---|---|---|---|---|---|---|
| S. aureus | 2.8 MBases | 131x | 2x 150bp | 15min | ~1GB | ~4GB |
| S. aureus | 2.8 MBases | 150x | 2x 250bp | 21min | ~1.6GB | ~6.5GB |
| E. coli | 5.5 MBases | 150x | 2x 150bp | 22min | ~2GB | ~8GB |
| E. coli | 5.5 MBases | 150x | 2x 250bp | 43min | ~5GB | ~20GB |
| P. aeruginosa | 6.2 MBases | 67x | 2x 150bp | 18min | ~2GB | ~8GB |
| P. aeruginosa | 6.2 MBases | 150x | 2x 250bp | 66min | ~8GB | ~32GB |
Note that actual time and memory requirements may be different depending on genome size, read length, coverage, read distribution, sequencing quality, and k-mer settings.
The table allows to estimate how many Velvet processes can be run on parallel. For example, on a system with 32GB, up to 4 Velvet processes can be run in parallel for a genome size of up to 6.2 MBases using downsampling to 150x and default clipping. For smaller genomes or less coverage, a higher number of processes can be run in parallel if enough processor cores are available. By default, SeqSphere+ starts one Velvet process per 8GB of RAM.
Open Source note
The Velvet assembler function is a wrapper to external Velvet executables. The Velvet software is open source software that is licensed under the GNU General Public License version 2.0 (GPLv2). The program homepage is http://www.ebi.ac.uk/~zerbino/velvet/. The program was ported to Windows by Applied Maths: http://www.applied-maths.com/download/open-source.
Please note that Ridom can only give limited support for the Velvet assembler.
For more information see: Zerbino DR and Birney E (2008) Velvet: algorithms for de novo short read assembly using de Bruijn graphs. Genome Res. 18(5):821-9 [PubMed 18349386].