Introduction
The E. coli CGE SerotypeFinder task template is based based on the CGE SerotypeFinder database (citation). The allele library contains nucleotide sequences for four O type genes (wzm, wzt, wzx, wzy) and five H type genes (fliC, flkA, fllA, flmA, flnA) that are searched by BLAST.
The task template can be downloaded from the Task Template Sphere (requires SeqSphere+ version 7.0 or later).
Task Entry Overview
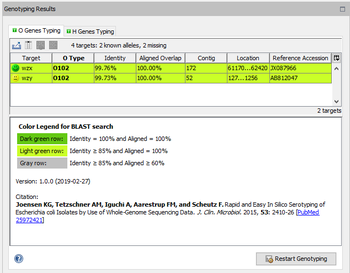
When an E. coli CGE SerotypeFinder task entry is processed, SeqSphere+ performs a target scanning for the defined O and H type alleles. The alleles that were found with at least 85% identity and 60% aligned overlap to the library allele are shown in the Task Entry Overview in two separate tables: 'H Type Genes' and 'O Type Genes'.
The rows in the two allele tables are colored by the percental identity and alignment overlap using the following thresholds:
- Dark green row: Identity = 100% and Aligned = 100%
- Light green row: Identity ≥ 85% and Aligned = 100%
- Gray row: Identity ≥ 85% and Aligned ≥ 60%
Below the table a colored threshold legend, version information, and citation(s) are stated.
Result Fields
For the confidently found (colored green) alleles the two fields 'O Type' and 'H Type' are rule based filled-in and 27 gene specific fields (e.g., 'Allele Type (wzm)', 'Reference Accession (wzm)', 'O Type (wzm)') are stored as result fields of the task entry.
The following rules are applied for fields 'O Type' and 'H Type' according to the publication by Joensen et al. (2015). J Clin Microbiol. 53: 2410-26:
Rules for result field 'O Type':
- If either wzx or wzy allele were found: If a result is given only for one gene then the unique result of this gene is used for the 'O Type' field.
- If wzx and wzy alleles were found: If one gene has a combination of types and the other gene has one of those types as unique result then the type of the unique result is used for the result field (this rule is not applied if one of the following types are involved: O13/O135 or O118/O151). Furthermore, no confident result is used if both genes are ambiguous (O17/O77/O44) or unique gene results contradict each other (O164/O124, O90/O127, O134/O46, or O162/O101).
- If wzx, wzy, wzm, and/or wzt were found: If the same unique type was found in all genes that have a result then this type is used for the 'O Type' field.
- In all other cases the 'O Type' result field is left empty.
Rules for result field 'H Type':
- fliC, fllA, flnA, flkA, flmA: If the same unique type was found in all genes that have a result then this type is used for the 'H Type' field.
- fliC and non-fliC (=fllA, flnA, flkA, flmA): If fliC has a different unique type than the unique type of the non-fliC genes then the non-fliC type is used for the 'H Type' field.
- In all other cases the 'H Type' result field is left empty.
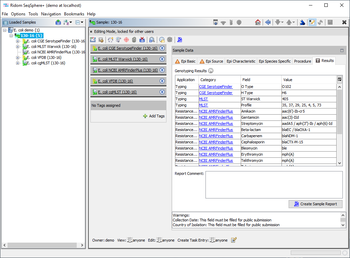
Only the two fields 'O Type' and 'H Type' are shown in the result tab of the Sample Overview. However, by clicking the CGE SerotypeFinder category links all details for this sample can be viewed.
All 29 result fields can be exported and selected from the CGE SerotypeFinder entry for searching under 'Field Criteria' in the advanced mode of the sample search dialog.
Only the two fields 'O Type' and 'H Type' are shown by default in a comparison table (with gray column header) when the task template was chosen. However, if needed the other 27 gene specific result fields can be added to a comparison table using the advanced settings.