Contents
Introduction
VFDB (citation) is a virulence factor database that is provided by the Institute of Pathogen Biology, Bejing, China. The database contains virulence factor related alleles for important bacterial pathogens.
The data at VFDB is only available for non-profit or authorized commercial users! (see terms of use)
Therefore, no predefined task templates can be provided. Non-profit or authorized commercial users can download the VFDB alleles manually and create an own local task template for a species, by using the steps below.
Citation
Chen L, Zheng D, Liu B, Yang J, and Jin Q. VFDB 2016: hierarchical and refined dataset for big data analysis--10 years on. Nucleic Acids Res. 2016, 44: D694-7 [PubMed 26578559]
Creating a VFDB Task Template
Downloading and Preparing VFDB Allele Libraries
- Step 1: Download from https://www.mgc.ac.cn/VFs/download.htm the file VFDB_setA_nt.fas.gz by clicking on DNA sequence of core dataset.
- Step 2: Start Ridom Typer and login.
- Step 3: Invoke in the menu “Tools > Gene Utilities > Split Sequence Library by Names” and open the FASTA file downloaded above.
- Step 4: Select the option Split by regular expression. Then copy following line and insert it into the regular expression field:
- (VFG\d+)(\((gb\|([\w\.]+))\))? \(([^\)]+\)?)\) (.*) \[([^\]]*)\] \[(\w+ \w+).*\]
- Step 5: Set the Fields(s) for subdirectory to 8, Fields(s) for library file to 5 and Field(s) for sequence name to 1,5,7. Choose for the delimiter to the “|”.
- Step 6: Confirm the dialog to export the files.
Creating a new VFDB Task Template for a Species
This describes how to create a new task temalate for a specific species, after downloading and preparing the allele libraries (see above)
- Step 1: Invoke in the menu “File > New > Create Task Template”
- Step 2: Click on “Create Task Template for Whole Genome Sequencing Data”
- Step 3: Click on “Create Task Template by Allele Libraries”.
- Step 4: Click on button “Import Allele Libraries”.
- Step 5: In the upcoming file browser go to the directory where the downloaded and prepared allele libraries are located. Double-click on the species directory for which a task template should be created. Then click on a FASTA file, press CTRL-A to select all FASTA files in the directory, and confirm the file dialog.
- Step 6: A preview of the allele libraries is shown. Leave all to defaults and confirm with OK.
- Step 7: A table shows the targets that will be created in this task template. All further settings should be left to defaults. Therefore confirm with Next two times, then click Finish to store the new task template
Updating an existing VFDB Task Template for a Species
This describes how to update an existing VFDB task temaplate for a specific species, after downloading and preparing the allele libraries (see above). The updating is possible for own created task template, and for the legacy VFDB task template downloaded from the Task Template Sphere.
- Step 1: Invoke in the menu “Options > Task Templates”
- Step 2: Select the VDFDB task template that should be updated and click the button "Update Allele Libraries" on the right.
- Step 3: In the upcoming file browser go to the directory where the downloaded and prepared allele libraries are located. Be sure to find the correct species directory for which this task template is defined and double-click it. Now click on a FASTA file, press CTRL-A to select all FASTA files in the directory, and confirm the file dialog.
- Step 4: A confirmation dialog with a preview of the update will be shown. It may be possible that due renaming some of the old targets do not exist any more. Existing samples will lose the results for those targets. If the number of removed targets is too high, be sure to check if the correct FASTA files were selected. When checked, confirm the dialog to start the updating.
After the updating has been performed, samples will not be updated automatically. The reprocess function can be used to restart the VFDB genotyping for them.
Task Entry Overview
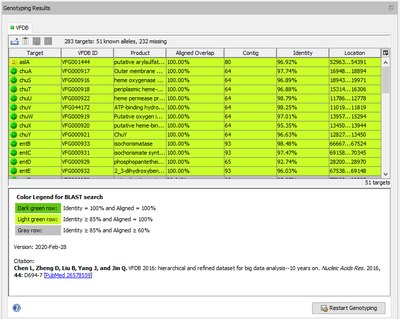
When a VFDB task entry is processed, Ridom Typer performs a target scanning for the defined virulence factor alleles. The alleles that were found with at least 85% identity and 60% aligned overlap to the allele in library are shown in the Task Entry Overview table. The rows in the table are colored by the percental identity and alignment overlap using the following thresholds:
- Dark green row: Identity = 100% and Aligned = 100%
- Light green row: Identity ≥ 85% and Aligned = 100%
- Gray row: Identity ≥ 85% and Aligned ≥ 60%
If multiple matches for a target (same or different allele) are found on different locations, each match is listed as separate row in the table.
Want to learn more about the virulence factor? Select the row of a virulence factor of interest, right-click, and choose the menu entry Browse VFDB. On the VFDB WWW page follow the link on top of the page for further information regarding this VF.
Below the table a colored threshold legend, version information, and citation(s) are stated.
Result Fields
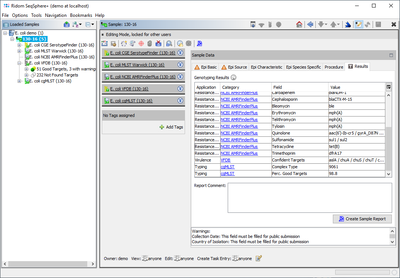
For each confidently found (colored green) virulence factor allele the Target name is stored as result field of the task entry (e.g., for 'aslA' = 'aslA'). If multiple matches for a target (same or different allele) are found on different locations, the gene appears multiple times concatenated with "," (e.g., for 'aslA' = 'aslA, aslA').
Additionally, the list of confidently found targets is stored in the result field 'Confident Targets' concatenated with "/". Only the latter summary result field is shown in the result tab of the Sample Overview. However, by clicking the VFDB category link all details for this sample can be viewed.
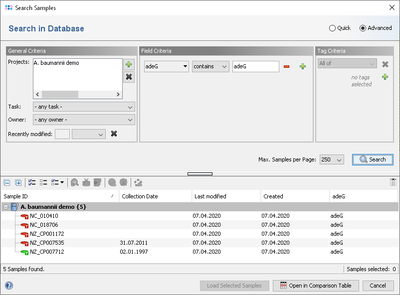
Specific Target fields can be selected from the VFDB entry for searching under 'Field Criteria' in the advanced mode of the sample search dialog.
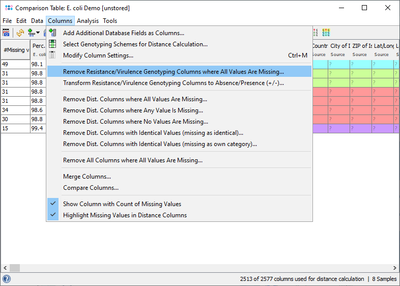
This result field can also be retrieved for a Comparison Table and for exporting metadata. If the VFDB Task Template is chosen in the Create Comparison Table dialog, then the Target data is shown right after the epidemiological metadata (with a gray column header). For a better overview it is recommended to use the command Columns | Remove Resistance/Virulence Genotyping Columns where All Values Are Missing to get rid of those columns that are for all samples empty.
If a virulence profile (presence/absence) comparison of several samples is intended to be done, then it is recommended to use the command Columns | Transform Resistance/Virulence Genotyping Columns to Absence/Presence (+/-). Alternatively handle missing values as an own category when building trees. Next the command ![]() Columns | Select Genotyping Schemes for Distance Calculation ... must be elicited and in the upcoming dialog VFDB must be selected and all other schemes should best be deselected from distance calculation. If data were not transformed, then once the command for calculating a tree was elicited in the upcoming missing values dialogue the option Missing Values are Own Category must be selected.
Columns | Select Genotyping Schemes for Distance Calculation ... must be elicited and in the upcoming dialog VFDB must be selected and all other schemes should best be deselected from distance calculation. If data were not transformed, then once the command for calculating a tree was elicited in the upcoming missing values dialogue the option Missing Values are Own Category must be selected.
Chromosome and Plasmids Overview
If the Chromosome and Plasmids Overview Task Template is used for the same Sample, some VFDB results are integrated there.
FOR RESEARCH USE ONLY. NOT FOR USE IN CLINICAL DIAGNOSTIC PROCEDURES.