Software Features
Client/server architectureThe client/server architecture allows distributed workgroups to cooperate via intra- or internet. Multiple users can access the data from various computers. Access rights and privileges can be configured to restrict access for specific users. | 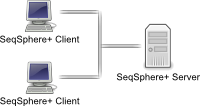 |
DNA re-sequencing editorAssemble raw reads (FASTQ) with integrated Velvet, SKESA, SPAdes, Flye*, or Raven* (latter combined with Medaka v2* and our proprietary ONT-cgMLST-Polisher*; JCM 63: in print, 2025) and BWA algorithms. Read and analyze assembly files (FASTA, GB, BAM, ACE). Inter-species contamination check with Mash Screen. Edit and analyze NGS or Sanger DNA sequence data. | 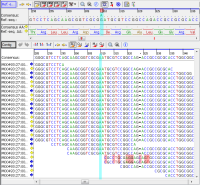 Editor for NGS data |
Pipeline for automated sequence analysisDefine and start a pipeline to trim, down-sample, assemble, analyze, and type hundreds of NGS data at once automatically. Define quality criteria for sample success. | |
Characterize bacteriaIdentify bacteria by using Mash Distance and FastANI. Type bacteria automatically by Genome-wide allele or SNP calling from WGS data either on core genome and/or accessory genome level. Use public schemes for genotyping or define them by using the incorporated cgMLST Target Definer. Use the NCBI AMRFinder for antimicrobial resistance (AMR) prediction. Apply Salmonella (SISTR), E. coli and L. monocytogenes geno-serotyping. MOB-suite and MobileElementFinder plasmid reconstruction and characterization. | 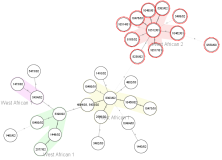 Minimum Spanning Tree |
DatabaseStore, search, retrieve, export, and create reports from experiment, circumstantial information, and DNA sequence data stored in an integrated database. Data fields are compliant with the meta-data requirements of the EBI European Nucleotide Archive (ENA). Compare new sequence entries against stored data. Get automatically clonal and/or plasmid transmission alerts* of clusters (Microbiol Spectr. 12: e0210024, 2024). Manage and backup all data (sequence and epi-data). Download from NCBI Genome complete/draft genomes or NCBI SRA reads. Submit with just one click raw read- and epi-data to EBI ENA. | 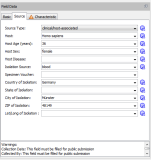 Editor for database fields |
Analytical toolsVisualize place, time, 'person', and type dimensions in a comparison table with built-in GIS, epi-curve, and phylogenetic tree (among others the minimum spanning tree algorithm is supported) functionality. All dimension views are interactively-linked and exportable in publication quality format (SVG and EMF). Store tree coloring, topology, and selection of samples in a comparison table snapshot. Visualize and compare plasmids with pyGenomeViz. Search for group-specific SNPs, e.g., for designing a cluster specific PCR screening assay (Microbiol Spectr. 10: e0303622, 2022 [PubMed]). | 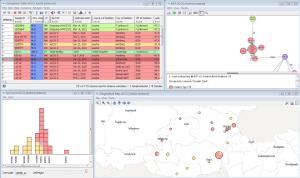 Place, time, 'person' and type dimension visualized |
CommunityDownload certified typing schemes online. Rapidly and easily share typing schemes with others. Contribute optional to the world-wide expanding publicly available cgMLST.org server of allele nomenclature. Consortium Server functionality allows several Ridom Typer users each with their own installation to share a minimum agreed upon amount of data. The shared data can be used for automatically triggered cross-institutional matching alert emails. | |
SecurityEncryption (SSL) of all data in transmission. Various configurable user roles, user groups and access controls. Audit trail functionality (who did, what, and when) for accredited laboratories. Configurable anonymization of meta-data like place and time before making data public (ENA submission). |
System Requirements
Microsoft Windows 10-(64bit), Windows 11 or Linux 64-bit, quad-core processor, 16-32 GB RAM, >1 TB disk (depending on the amount of data that should be stored). See the Ridom Typer documentation for more detailed system requirements.
Licensing
The license is limited to a period and a certain number of named user accounts on the server. During the license period an unlimited number of samples can be analyzed. Each named user account allows to login interactively once at the same time. If somebody else logs in with the same account, a read-only mode is activated for this session. The Pipeline Mode also runs on behalf of a named user account but can run concurrently multiple times, i.e., for a 2 named user license in total 5, for 5 users in total 12, and for 30 users in total 75 concurrent pipelines are possible, respectively. Client software can be installed on an unlimited number of computers. The license period starts with the activation of the license. The license will expire after the defined period and the software runs with read-only access unless a new license is ordered. Read-only access still allows to view and export existing data. Licenses include updates of Bruker MBioSEQ Ridom Typer and the purchased modules and free technical support for installation and troubleshooting of the software within the period.
* Part of the extra charged Long Read Module (LRM) that includes the ONT Data Assembly and Long Read Data Plasmid Transmission Analysis sub-modules.
FOR RESEARCH USE ONLY. NOT FOR USE IN CLINICAL DIAGNOSTIC PROCEDURES.