Introduction
The CGE MobileElementFinder task template uses the tool MobileElementFinder for identifying integrative Mobile Genetic Elements (iMGEs) in WGS sequence data [PubMed 33009809].
The tool is designed for rapid detection of iMGEs and their genetic context. The detection is based on sequence similarity to a database of known elements and a prediction of putative composite transposons. Data were compiled from the ISfinder, Transposon Registry, and ICEberg databases. The tool's ability to predict novel iMGEs is limited. Furthermore, it can not handle circularity information. Putative composite transposons are flagged if there are two IS of the same type located on the same contig where the total length of the sequences and the intermediary region is less than 52.5 kb.
Abbreviations used by MobileElementFinder:
- IS - insertion sequence
- cn - composite transposon (named after length of intermediary region and defining ISs, e.g., cn_12127_IS26)
- Tn - (unit) transposon
- MITE - miniature inverted repeat
- ICE - integrative conjugative element
- CIME - cis-mobilizable element
- IME - integrative mobilizable element
![]() Important:
Important:
- If Ridom Typer is installed on Windows, MobileElementFinder requires the Windows Subsystem for Linux (WSL).
- If Ridom Typer is installed on Linux, MobileElementFinder must once be installed by calling the installation of Bioinformatic Tools on Linux.
Task Entry Overview
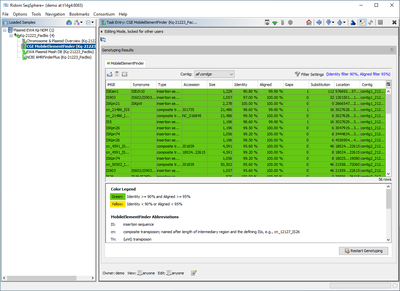
The Task Entry Overview of the processed task entry shows the MobileElementFinder result in a table. In the Task Template it can be configured to ignore Samples with more than 30 contigs, i.e., short-read data (by default off). The table is by default sorted according to contig and location. It is possible to filter for a certain contig only. The table rows are colored by the percental identity and alignment overlap using the following thresholds:
- Green row: Identity ≥ 90% and Aligned Overlap ≥ 95%
- Yellow row: Identity < 90% or Aligned Overlap < 95%
By default, the table is filtered to show only the reliable ("green") matches. The MobileElementFinder report table contains the following columns (but only bold columns are shown by default):
- Number: A reference number for the found iMGE, or a combination of multiple numbers for composite transposons
- iMGE: The name of the found iMGE, or a combination for composite transposons
- Synonyms: Synonyms of the name
- Type: Type of the iMGE (e.g., 'composite transposon')
- Accession: Reference accession number
- Size: Size of the iMGE reference
- Identity: Percental identity of BLAST match
- Aligned: Percental aligned overlap of BLAST match
- Gaps: Gaps in BLAST alignemnt
- Substitution: Substitutions in BLAST alignment
- Location: Location of BLAST match on contig
- Contig: Contig name
- Start: Start of BLAST match on contig
- End: End of BLAST match on contig
- Cigar: BLAST alignment CIGAR string
- Depth: Always empty, as only contigs are queried by Ridom Typer
- E-value: E-value of BLAST match
- Prediction: Contains 'predicted' or 'putative'
The toolbar button ![]() can be used to export the iMGE sequences for selected rows.
Below the table a colored threshold legend, version information, and citation(s) are stated.
can be used to export the iMGE sequences for selected rows.
Below the table a colored threshold legend, version information, and citation(s) are stated.
Result Fields
The task entry has only one result field, that stores the number of iMGEs that were found:
- iMGEs Found
Chromosome and Plasmids Overview
If the Chromosome and Plasmids Overview Task Template is used for the same Sample, the MobileElementFinder results are integrated there.
FOR RESEARCH USE ONLY. NOT FOR USE IN CLINICAL DIAGNOSTIC PROCEDURES.