Contents
Overview
Early Warning Alerts (EWA) are used to automatically detect, when a newly processed sample or its plasmids (query) are similar to samples or plasmids that are already in the database (hits). For samples, similarity is assessed based on cgMLST allelic profiles and for plasmids it is based on Mash distances. If similar samples or plasmids within the defined similarity thresholds are found, an EWA is triggered and stored in the database.
Please Note: Using Plasmid Early Warning Alerts requires the Long Read Module (LRM)
Defining cgMLST Early Warning Alerts
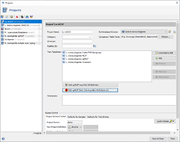
The cgMLST Early Warning Alert definitions can be created and managed by pressing the button Add cgMLST Early Warning Alert Definition in the project editor below the task templates section. The button is only enabled if a cgMLST Task Template was added to the project. The button icon is grey if no EW-Alerts are defined, else it is colored red.
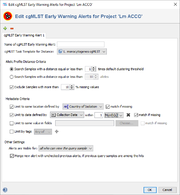
An EWA definition contains the following sections:
- Name of cgMLST Early Warning Alert Definition
- A name that can be used as a reference for this EWA definition. The name is shown in the alerts that are triggered. If only one EWA is defined for a project, the name can be left empty.
- cgMLST Task Template for Distance
- If the projects contains multiple cgMLST Task Templates the one to be used for distance calculation can be selected here.
- Allelic Profile Distance Criteria
- Defines the threshold for similar samples. By default, the clustering threshold of the cgMLST Task Template is used. Alternatively a multiple of the clustering threshold, an absolute allele distance, or with a similarity above percentage threshold can be chosen.
- A filter allows to exclude samples with too much missing values from the search. By default all samples with more than 10% missing values will be excluded.
- If the S. aureus spa-typing task template is chosen for distance calculation, the section is replaced a Spa-Type Distance Criteria section. The alert is than triggered if samples with the same spa-type are found.
- Metadata Criteria
- Allows to define metadata limit criteria for the alert. If multiple limits are defined they will be linked by the AND operator.
- Limit to same location: The hit sample must have the same location as the query sample. If this limit is used the option match if missing is set by default. Thereby an alert is triggered even if the query and/or hit sample location field is empty. If this option is deselected no alert is reported if the query and/or hit sample location field is empty. The Country of Isolation or the City of Isolation can be defined as location field.
- Limit to date: The date of the hit sample must match within a given time period (days, weeks, months, or years) with the date of the query sample. If this limit is used the option match if missing is set by default. Thereby an alert is triggered even if the query and/or hit sample date field is empty. The Collection Date or Created for the sample entry creation date can be defined as date field.
- Limit to same value in fields: Other database fields than location and/or date fields that must match can be selected here. If this limit is used the option match if missing is deselected by default. Thereby no alert is reported if the query and/or hit user defined fields is empty.
- Limit by tags: Only query samples with the defined tag(s) will initiate the alert search and only similar samples that fulfill the tag criteria will be reported as hit samples. If multiple tags are selected they will be linked by the AND operator.
 Hint: The EWA searches are performed directly after a new sample is processed and stored. Therefore, the metadata (location, date, user defined fields, and tags) must be entered in advance (e.g., by spreadsheet import of metadata, pipeline tags, and/or spec files) if they should be effective as limiting criteria in the EWA.
Hint: The EWA searches are performed directly after a new sample is processed and stored. Therefore, the metadata (location, date, user defined fields, and tags) must be entered in advance (e.g., by spreadsheet import of metadata, pipeline tags, and/or spec files) if they should be effective as limiting criteria in the EWA.
- Other Settings
- By default, alerts are visible for all users that can view the query sample. However, the definition dialog allows to limit the visibility of the triggered alerts to a specific user group. Users who don't belong to this group will not see the alert. The alert is always visible for the user who imported the sample that triggered the alert. If the alert is visible for multiple users, all of those users can mark the alert as checked or delete the alert. This alert status applies then to all users who can view the alert.
- If the Merge new alert ... option is enabled (default), new alerts are merged with unchecked older alerts, if the query sample of an unchecked previous alert is among the hits of a newer alert, i.e., the new alert gets the query sample and all hit samples of the old alert assigned and the old alert is deleted. Please note that only unchecked alarms are merged into new alerts. Thereby, the number of triggered alerts (e.g., from one large outbreak) can be reduced effectively.
By pressing the buttons OK of the Edit cgMLST EWA definitions dialog and the Save & Close of the project window the EWA is stored and active.
Adding and Deleting cgMLST Early Warning Alerts
EWAs can be added and deleted at any time when in the project manger window a project is selected and the Edit cgMLST Early Warning Alert Definitions button of this project is pushed. For adding a EWA the ![]() icon must be pushed and the new EWA must be defined. For deleting an EWA the corresponding tab must be selected and the
icon must be pushed and the new EWA must be defined. For deleting an EWA the corresponding tab must be selected and the ![]() icon must be pushed. Once the buttons OK of the Edit cgMLST Early Warning Alert Definitions dialog and Save & Close of the project dialog are clicked the definition is added or deleted.
icon must be pushed. Once the buttons OK of the Edit cgMLST Early Warning Alert Definitions dialog and Save & Close of the project dialog are clicked the definition is added or deleted.
Defining Plasmid Early Warning Alerts
Please Note: Using Plasmid Early Warning Alerts requires the Long Read Module (LRM)
Unlike the EWAs for cgMLST, the plasmid EWAs are defined for a Typing, and not for a Project.
The Plasmid Early Warning Alert definitions can be created and managed using the menu command Options > Mash Plasmid Databases. The EWAs can be modified using the button ![]() Edit Early Warning Alert.
Edit Early Warning Alert.
See the page Manage Plasmid Databases for details. The Metadata Criteria work the same way as described in the section for cgMLST Early Warning Alerts above.
Performing cgMLST Early Warning Alert Searches
If Early Warning Alert(s) are defined for a project, the EWA search(es) are automatically performed when a sample is added to this project via either the Pipeline Mode or the command Process Assembled Genome Data.
Performing Plasmid Early Warning Alert Searches
Please Note: Using Plasmid Early Warning Alerts requires the Long Read Module (LRM)
If plasmid EWA search(es) are enabled, they are automatically performed when a plasmid typing is done on a sample that is imported via the Pipeline Mode. Plasmid Transmission EWAs are triggered if a Sample that is processed by the pipeline using the plasmid typing has plasmids with a Mash distance below the distance threshold to another plasmid in the database.
If a Sample contains more than one plasmid, multiple EWAs can be created for this Sample, one for each plasmid. However, to prevent plasmid alerts for clonal transmissions, no alert is reported for a plasmid hit if the respective Samples have a cgMLST distance below the specified cluster threshold. If more than one cgMLST typing is present in a Sample, only the cgMLST typing scheme that is listed first in the project definition will be considered.
Note: The client that runs the pipeline requires the Linux tools to trigger a plasmid EWA.
Managing Triggered Early Warning Alerts
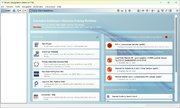
The three most recent and unchecked clonal and plasmid EWAs that were triggered are shown on the top of the home screen in the section Unchecked Early Warning Alerts. If an EWA is opened by clicking on it, a plasmid table with the involved plasmids or a comparison table with the involved samples is shown.
Please Note: Using Plasmid Early Warning alerts requires the Long Read Module (LRM)
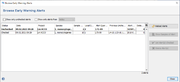
In the Options main menu the command Browse Early Warning Alerts ... must be issued to list all triggered EWAs that are visible for the user in a table.
The column Transmission describes the transmission type:
Plasmid EWAs are marked by ![]() Plasmid, cgMLST EWAs by
Plasmid, cgMLST EWAs by ![]() Clonal.
Clonal.
By default the alerts are sorted according to the triggering date. The table can be filtered for unchecked alerts only (default) and/or for a time period. The button panel on the right (or the right-click menu) can be used to set multiple alerts as checked or to delete them. Additionally, a comparison table/a plasmid table can be opened by pushing the button Show Samples of Alert (or by double-clicking in a row) for a selected alert containing all samples or plasmids that are involved in the alert.
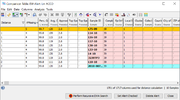
cgMLST Early Warning Comparison Table
The query sample, the former query samples of merged alerts, and the hit samples are grouped by colors in the comparison table.
On the bottom of the comparison table window three EWA specific buttons are available:
- Perform Recursive EWA Search
- In the inital EWA search only hit samples are found, that are within the defined allelic threshold compared to the query sample. If a recursive EWA search is performed, all samples that are within the defined allelic threshold to any hit samples are returned also. This is repeated until no additional hit samples are found. The recursive EWA search applies also all limiting metadata criteria that were used for the original EWA search.
- Set Alert Checked
- Sets an unchecked alert to checked. The alert is automatically removed from the home screen but is still available in the database. A checked alert will not any longer be merged with any newer alert(s).
- Delete Alert
- Deletes the alert completely from the database.
 Hint: If it is intended to increase the discriminatory power for the involved samples of an EWA cluster, the accessory genome targets could be added for distance calculation in the comparison table. Issue the command Columns | Select Genotyping Scheme for Distance Calculation ... and select in the upcoming dialog the Accessory Task Template in addition to the cgMLST Task Template.
Hint: If it is intended to increase the discriminatory power for the involved samples of an EWA cluster, the accessory genome targets could be added for distance calculation in the comparison table. Issue the command Columns | Select Genotyping Scheme for Distance Calculation ... and select in the upcoming dialog the Accessory Task Template in addition to the cgMLST Task Template.
FOR RESEARCH USE ONLY. NOT FOR USE IN CLINICAL DIAGNOSTIC PROCEDURES.