The workflow of a pipeline is defined by a pipeline script. Pipeline scripts are always stored on the computer in the profile directory of the user where the Ridom Typer Client is installed that was used to define a script. To share a script on multiple Client computers the script can be either ex- and imported or distributed via pipeline reports. A pipeline script can be easily created using the wizard dialog which has the following six sections.
Contents
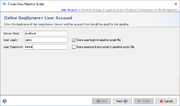
User Account
To setup a pipeline script a user account on a specific server is required. By default, the User Login but not the User Password will be stored in a pipeline script. Thereby the script can only be edited and run when the Password of this user is entered. This means the pipeline will be run on behalf of this user, and all Samples created by such a defined pipeline script will be owned by this user.
If the User Login is not stored in the script, a login dialog is shown when starting the pipeline and the pipeline runs with the supplied login information.
Next to the User Login also the User Password of the user can be stored in a pipeline script. The password is always stored encrypted. Every user can run such a script. Export such a script to share it between different users. However, for editing a script always the User Password is required.
![]() Warning: If User Login and User Password are stored in the pipeline script, then anyone who has access to the computer account can run a pipeline without further authentication!
Warning: If User Login and User Password are stored in the pipeline script, then anyone who has access to the computer account can run a pipeline without further authentication!
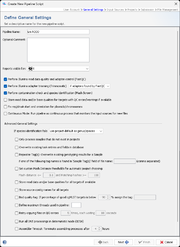
General Settings
The section defines general settings of the pipeline. Each pipeline must have a unique name. Optionally a comment can be entered. In addition the access rights for the reports of the pipeline can be specified here (default is primary group).
The following options can be selected:
- Perform read data quality and Illumina adapter control (FastQC)
- This by default enabled option can be used to perform read data quality and adapter control with FastQC.
- Perform Illumina adapter trimming (Trimmomatic)
- This by default enabled option can be used to perform Illumina adapter trimming with Trimmomatic. It contains two values of always in which Trimmomatic will be executed all the time before FastQC step. The other option is if adapters found by FastQC(default) that performs adapter trimming only if the FastQC result status be reported as warning or failed. Consequently it will be executed only if FastQC perform. In following, FastQC will be executed once again on trimmed read data.
- Perform contamination check and species identification (Mash Screen)
- When enabled, Mash Screen is performed against a reference database to find contamination with other species above a ratio of 10%. The top match is also used to set the Genus and Species field of the Sample (enabled by default for new pipeline scripts if available).
- Store read data and/or base qualities for targets with QC errors/warnings if available
- This can be used to define that the read data should be kept for targets with QC errors or warnings. By default they are not kept. However, QC messages (e.g., low coverage) are still preserved.
- Fix rep/dnaA start and orientation for plasmids/chromosomes
- This by default enabled option is used to automatically perform the Fix Start and Orientation for Plasmids or Chromosomes function on circular contigs, i.e. the options look for [topology=circular] and skip non-circular contigs are enabled for reorientation. This function will only be performed in importing pipelines, not for pipelines that are reprocessing samples from the database.
Please Note: Fix start and orientation requires the Long Read Module (LRM)
- Continuous Mode: Run pipeline as continuous process that monitors the input source for new files
- This can be used to define a pipeline that waits for new files in a specific directory. When started, the pipeline processes all existing files found in the input sources. After the files are processed, the pipeline waits until new files appear in the input source(s) and then starts again processing the new files (the pipeline checks every 10s for new files, but checks that new files are older than 1min, to assure that they are not written currently).
By pressing the Advanced General Settings button more advanced options can be selected:
- If species identification fails
- Defines how to set Genus and Species of the sample, if the species cannot be identified by Mash (e.g., for closely related species).
- Only process samples that do not exist in projects
- If this option is enabled, only files for those samples that do not already exist in one of the projects used in the pipeline are processed. For large input directories and projects this speeds up the skipping of files that were already processed, especially if multiple pipelines are started in parallele.
- Overwrite existing task entries of samples in database
- By default, files are only processed if they belong to new samples or provide new data (new FASTQs and/or task(s)) for existing samples, according to the Sample ID configured in the File Naming. If this option is enabled, any task entry in an existing sample will be overwritten by the pipeline (e.g., FASTQs should be assembled with another assembler). Important notice: A pipeline script with this option enabled should not be run in parallel pipelines. If such a script is run in parallel pipelines, the samples are processed once per pipeline!
- Repeater Tag(s)
- This feature can be used to handle repeatedly sequenced samples in a controlled manner. The tag name that is defined here must be a part with delimiters of the FASTQ file names of the repeatedly sequenced samples. The position of the tag in the file name must be stated in the Sample Tag of the File Naming definition section of the script. If multiple repeated runs are done, a list of comma separated tag names can be entered here. If in a pipeline script repeater tag(s) are defined then the default mechanism of doing a "merged assembly" is turned off. Please see the FAQ for more details.
- By default, the repeater tag feature is disabled and from repeatedly sequenced samples a "merged assembly" is done by default automatically.
- Set custom Mash Distance thresholds for automatic project choosing
- This can be used to overwrite the default thresholds for Mash-distance (default <=0.1) and Matching-hashes (>=100) that are used in the automatic project choosing (Mash Distance).
- Store read data and/or base qualities for all targets
- This can be used to define that the read data should be kept for all targets. By default they are only kept for targets with QC errors or warnings.
- Store source contig names for all targets
- This can be used to define that the original contig names should be kept for all targets.
- Bad quality tag
- This can be used to define a name for a tag that is automatically assigned to a Sample, if the percentage of good cgMLST targets is below the given value. If no cgMLST Task Template is used, this parameter has no effect. A tag helps to search for specific Samples (e.g., all bad quality tag Samples since yesterday) more efficiently in the database.
- Define maximum threads used in pipeline
- This can be used to set default maximum thread count and it overrides the maximum available threads. Also this figure can be overridden by advanced assembler configurations.
- Retry copying files on I/O errors
- Allows to specify to retry copying files (e.g., FASTQs) if temporary disk problems occur when storing the assembly files, e.g. because of temporarily unavailable network drives.
- Run all ONT processings in deterministic mode (BETA)
- Chopper, Rasusa, and Flye have non-deterministic steps in their processing. To a assure that the processings run as deterministic as possible, this option can be set. However, this increases the runtime dramatically and is therefore only recommended for evaluation projects!
- Assembler Timeout
- This can be used to terminate an assembling/mapping processes if it takes too long.
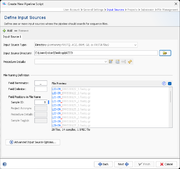
Input Sources
An input source defines where the pipeline should look for sequencing data (FASTQ/ACE/BAM/GB/FASTA files). FASTQ files with adapters (& multiplex indices) trimmed-off are required here for optimal de novo assembly results. Multiple input sources can be defined to process different samples from different directories and sequencing machine vendors (e.g., Illumina or Oxford Nanopore) by pushing the + Add button in the upper-left corner.
One of the following Input Source Types can be chosen:
- Directory
- the directory on a local or remote (e.g., CIFS, NFS) disk defined in the 'Input Source Directory' field.
- Directory and subdirectories
- the directory disk defined in the 'Input Source Directory' field including all subdirectories.
- iSeq/MiniSeqMiSeq/NextSeq hard disk
- the disk of the Illumina iSeq/MiniSeqMiSeq/NextSeq or the run folders copied to a network drive, location disk defined in the 'Input Source Directory' field.
- Oxford Nanopore MinKNOW hard disk
- the working directory or a run directory of Oxford Nanopore MinKNOW run data.
- When Oxford Nanopore MinKNOW hard disk is selected as Input Source Type an additional input field is available: Output FASTQ directory.
- The FASTQ-files for a MinKNOW sample are merged together and stored in a subdirectory of Output FASTQ directory that is named by the run id. Therefore this directory must be writeable for the current user.
- Note that Field Terminator and Field Delimiter cannot be edited when Oxford Nanopore MinKnow hard disk is selected and that the file preview is disabled.
- From Samples in Database
- the assembly contigs of all samples of the projects selected in the pipeline script are used as input. The pipeline can be optionally limited to samples that do or do not contain specific tags. This input source type allows to reprocess tasks or process new tasks more efficiently for a large number of samples.
The sequencing vendor (e.g., Illumina or Oxford Nanopore, etc.) and sequencing protocol (e.g., single-end or paired-end reads) of the Procedure Details must be defined always if FASTQ files will be assembled and no other laboratory procedure details are available (e.g., by file naming procedure details or by SPEC files). All other procedure details are optional. However, if it is intended to submit FASTQ files to ENA then please remember that some laboratory procedure details are obligatory required by ENA. If assembled data is processed with the pipeline (ACE/BAM/GB/FASTA files) then procedure details are always optionally. In general this feature is very convenient way to attach detailed - frequently identical - laboratory procedure details for documentation purposes to a large number of Samples.
File Naming
Each input source must have a sequence data file naming convention defined, that enables an automatized workflow with no user intervention required. Minimum the Sample ID information must be transferred by the sequence data file name. However, additional information (in file name fields) can be transferred via the file name to further streamline this automized analysis. Usually the file names of sequence data can be controlled to a certain amount by supplying a filled-in sample sheet (e.g., by using the Illumina Experiment Manager tool) to a sequencing machine before starting a run.
First, the file name can be split into different fields by a Field Delimiter until a defined Field Terminator appears. If no terminator is defined, the whole file name (without file extension) is used.
Next the Field Positions in File Name of the various fields can be defined here, by entering a number (beginning with 1) into
- Sample ID
- Defines the position(s) of one or more tag(s) fields in the file name that used as Sample ID (only required information). Multiple positions can be separated with commas here and will be concatenated with the field delimiter defined above. If in the pipeline project no Sample with this ID exists then the pipeline creates a new Sample entry with this ID and attaches the genotyping results from the sequence data to this entry. If a Sample with this ID and with associated epi metadata but no genotyping results already exists in the pipeline project then the pipeline attaches the genotyping results from the sequence data to this entry. If a Sample with this ID and associated genotyping results for the foreseen Task Templates already exists in the pipeline project, and if the pipeline does not provide new FASTQ files for this Sample, then the Sample is not processed at all. If the pipeline provides new FASTQ files (same Sample ID but different file name than previous FASTQ, i.e., repeatedly sequenced sample) for this Sample, and if the old FASTQ files of the Sample are still available at the stored location, then by default the old and the new FASTQ files are assembled together, and the Sample is updated with the new assembly and genotyping result. Please see the FAQ for more details about repeatedly sequenced samples.
- Project Acronym
- Defines the position of the project acronym field in the file name. If an acronym field is defined here, the matching acronym for each sample will be searched in the defined projects of the pipeline script. The acronym field can be left empty if Samples from only one project are going to be processed or if the automatic project choosing by Mash Distance is used.
- Procedure Details
- Defines the position of the procedure details name field in the file name (can be left empty). If this feature is used in sequence data files and defined here then the associated Sample entries get the Procedure Details field values entered of the object that is referred to by the name. If this option is used it overwrites where applicable also supplied SPEC file information and always the procedure details stated above in this section.
- Sample Tag(s)
- Defines the position(s) of one or more tag(s) fields in the file name (can be left empty). Multiple tag positions can be separated with commas here. If this feature is used in sequence data files and defined here then the associated Sample entries get the tag(s) attached to them. Next those tag(s) can be used to search for Samples more specifically in the database. The position of repeater tag(s) in the file name need also to be defined here.
The coloring of the sequence data files in the File preview box documents how current delimiter, terminator, and field settings apply to those file.
![]() Hint: Illumina uses the '_' as a separator in file names. Therefore, it cannot be used as field delimiter for the file naming. We recommend to use '-' as field delimiter and '_' as field terminator for Illumina FASTQ files.
Hint: Illumina uses the '_' as a separator in file names. Therefore, it cannot be used as field delimiter for the file naming. We recommend to use '-' as field delimiter and '_' as field terminator for Illumina FASTQ files.
Advanced Input Source Options
- Append to Sample IDs
- The field can be used to append a text automatically to the Sample IDs that are imported from this input source. This can be used for comparing the same sample with difference processing options or Ridom Typer versions. The text can contain also the following place holders that will be replaces:
%twill be replaced with the current timestamp when processing the sample%vwill be replaced with the Ridom Typer version%s1...%s9will be replaced with the 1st ... 9th word of the pipeline name (words are separated by blank)
- Default Project Acronym
- If a project acronym field is not used in all sequence file names, a default value can be entered here, that will be used for Samples with no associated project information that are imported from this input source. This field can also be used to assign a input source to a specific project if no acronyms are used in the sequence file names.
- Assign Tag(s)
- If no tag information is transferred by (all) sequence data file names it might be in some cases useful to assign tag(s) to all Samples that are imported from this input source. Multiple tags can be attached for which the names must be entered separated by comma here.
- Use Regular Expression
- The file naming can also be defined by a regular expression that splits the file name into various fields. Once this option is chosen and a regular expression is entered the 'Field Delimiter' and 'Field Terminator ' entered above are ignored.
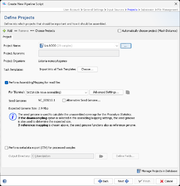
Projects
In the project section it is defined into what projects Sample data will be imported. At least one project must be defined here even if a project acronym is used in the file names and its position is defined in the previous section. If more than one project is defined here, automatic project choosing must be enabled, or each project must have an acronym that is used in the file names and their position defined in the previous section. In this case the project for each Sample is selected by the acronym that is found in the file name of the input data.
If the checkbox Automatically choose project (Mash Distance) is selected, the pipeline uses Mash Distance to choose the project for a Sample out of the list of projects defined in the pipeline script. Therefore, only one project per species can be defined in the pipeline if this option is used. The option is disabled if project acronyms are used.
By pushing the ![]() Add or
Add or ![]() Remove buttons in the upper-left corner the projects to be used by the pipeline script can be managed.
Or use the
Remove buttons in the upper-left corner the projects to be used by the pipeline script can be managed.
Or use the ![]() Choose buttons to directly select multiple projects.
The Manage Projects in Database button in the lower-right corner of this section can be used to modify existing projects (e.g., adding a project acronym) or to create new projects.
Choose buttons to directly select multiple projects.
The Manage Projects in Database button in the lower-right corner of this section can be used to modify existing projects (e.g., adding a project acronym) or to create new projects.
Per project tab the following information needs to be selected or is shown:
- Project Name
- Via a drop-down list a project can be chosen. By pushing the ... button the project list can be filtered.
- Project Acronym (not editable)
- Shows the project acronym (if defined at all) for the chosen project.
- Project Organism (not editable)
- Shows the project organism as entered in the Task Template(s) for the chosen project.
- Task Templates
- By default all Task Templates of a selected project are chosen by a pipeline script. By pushing the Choose... button next to Task Templates this can be limited to selected Task Templates.
The checkbox Perform Assembling/Mapping for read files can also be checked and configured per project. Once selected a pipeline using this script will assemble FASTQ files (checkbox obviously does not apply for ACE/BAM/GB/FASTa files). For each sequencing vendor (defined in the input sources section via multiple input sources and/or by using a procedure details field in the FASTQ file names) an assembler or mapper can be selected. The following assemblers/mappers are available:
- SKESA short-read de novo assembler (recommended only for Illumina data; only available for Linux or WSL)
- SPAdes short-read de novo assembler (only available for Linux or WSL 2)
- Velvet short-read de novo assembler (only for Illumina data; legacy, only available for Windows)
- BWA short-read reference mapper
- Flye long-read de novo assembler (recommended for Oxford Nanopore data; only available for Linux or WSL)1)
- Raven long-read de novo assembler (recommended for Oxford Nanopore data; only available for Linux or WSL)1)
1) Only available with ONT Data Assembly Module
Right to the assemblers/mappers drop-down selection list, an Advanced Settings button can be used to specify the preprocessing (trimming, downsampling) and assembler/mapper parameters. For short read data downsampling is selected here by default for Velvet, SKESA, SPAdes, and BWA. Trimming is only turned on by default for Velvet and can again be modified in the Advanced Settings. By default remapping of reads and consensus sequence polishing is turned on for SKESA and SPAdes (for Velvet this is not an option as this is obligatory). Velvet and BWA produce always a ACE or BAM file, respectively. With default parameters SKESA and SPAdes produce BAM files else a FASTA assembly contig file only that is always produced by all assemblers. For long read data downsampling, trimming and Medaka polishing is by default enabled for Flye and Raven assembling. Both assembling methods produce always a FASTA file only.
If multiple projects are chosen in the pipeline, the second and further ones are by default configured to use the same assembler and assembler settings as the first project. This can be changed by selecting an assembler in the drop-down list.
The project Seed Genome and Expected Genome Size are shown below the selected assembler/mapper. A seed genome is required if reference mapping (BWA) is chosen, or if the downsampling option is selected in the Advanced Settings. The seed genome is also used to calculate the unassembled coverage for the Procedure Statistics. If the Project contains a cgMLST Task Template then the seed genome is automatically taken from the Task Template (if multiple cgMLST Task Templates should be present in a single project then the seed genome of the Task Template first in ordering is taken). The seed genome can also be set manually by using the Alternative Seed Genome button. In the case that no cgMLST Task Template is present in the project then this button must always be used to define a seed genome. For multi-species projects, or if no cgMLST is available, the option Find Expected Genome Size automatically (Mash Distance) can be used to determine the genome size by a close matching reference genome.
Additionally, the use of MOB-recon for the Chromosome and Plasmids Overview Task Template can be enabled/disabled using the checkbox Perform reconstruction of chromosome & plasmids (MOB-recon). If MOB-recon is disabled, contigs shorther than 500.000 bases are assumed to be a closed plasmid (e.g. PacBio data) and are directly passed to MOB-typer.
At the bottom of the panel the Export metadata option can be enabled to export for each sample that is processed in the pipeline a CSV file containing metadata fields. The set of fields that will be exported can be configured or can be left to the project default fields. Thereby, Ridom Typer results can be automatically imported in other tools (e.g., for making a customized report).
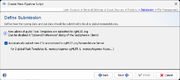
Submission
For public cgMLST Task Templates the option Automatically submit new CTs anonymized to cgMLST.org Nomenclature Server can be enabled here. If a sample does not belong to an existing Complex Type (CT), it is required to submit the allelic profile to define a new CT. The allelic profile of the new CT will then be stored on cgMLST.org. No epi data or submitter data of the sample will be stored on cgMLST.org. Submission to cgMLST.org is only possible if the Sample has in the cgMLST scheme at least 90% of good targets (i.e., targets passed the QC procedure and have therefore a green or yellow smiley).
For local Task Templates the default setting is to automatically assign new types (if this option is enabled in the Task Template).
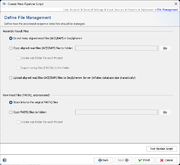
File Management
In this last section it can be defined how the pipeline process should manage the assembled/mapped files (ACE/BAM) and the read files (FASTQ). All files are referenced in the process tab of a Sample (see more details).
The are three options how to handle Assembled/Mapped Files (ACE/BAM):
- Do not keep ACE/BAM files in Ridom Typer
- This is the default and stores with the Sample only a link to the ACE/BAM file. If the file path of the link is referring to a local disk of a specific computer that has the Ridom Typer client installed, than the link is only working when used by a Ridom Typer client from exactly this computer. Furthermore, if this option is used in a pipeline that performs assembling/mapping with ACE/BAM result files (processing of FASTQs with Velvet or BWA produce always ACE or BAM files, respectively; when using SKESA and SPAdes with default parameters BAM files are created due to the 'polishing' step) then those files will not be kept permanently and the links to them will be broken.
- Copy ACE/BAM files to folder
- If this option is chosen, ACE/BAM files are copied to the selected directory. The Sample entry stores only a link to this copied file. Subdirectories named by the project acronym (if available) with the according ACE/BAM files can be created automatically when the Create sub-folder for each Project option is chosen. When the option Additionally write contigs to FASTA files is chosen then next to the ACE/BAM files the FASTA assembly contigs files are also exported into the according (sub)folder.
- Upload files to Ridom Typer Server
- If this option is used, the assembled/mapped files are uploaded to and stored at the Ridom Typer Server. The uploaded file is automatically deleted when the attachment is removed in the process tab of the Sample or if the whole Sample is deleted. Using this option will dramatically increase the databases size. Therefore, it is not recommended to use this option permanently in pipelines.
The very large Raw Read Files (FASTQ) cannot be uploaded to the Ridom Typer Server. By default the option Store links to the original FASTQ files is turned on that stores the path to FASTQ(s) in the procedure tab of the Sample entry. However, again there is an option to Copy FASTQ files to folder that copies the files to a selected folder. The Sample entry stores only a link to this copied file. Subdirectories named by the project acronym (if available) with the according FASTQ files can be created automatically when the Create sub-folder for each Project option is chosen.
FOR RESEARCH USE ONLY. NOT FOR USE IN CLINICAL DIAGNOSTIC PROCEDURES.