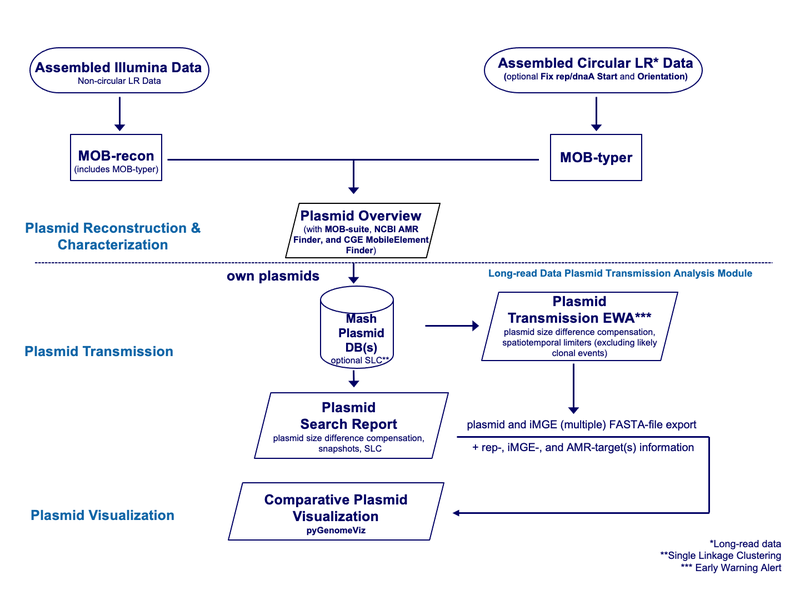
Overview
The basis for this module is the usage of the Chromosome and Plasmids Overview (MOB-suite), NCBI AMRFinderPlus, and CGE MobileElementFinder task templates that are available for all users. The module then enables users to detect plasmid transmissions using the Mash Plasmid Typing function. Therefore, own Mash Plasmid databases can be created. These databases can be created for all plasmids of a sample or only plasmids that contain certain AMR targets. Optionally, a single linkage clustering of the plasmids in the database can be performed. Plasmid searches can be used to compare a single plasmid with the database content and find similar plasmids using a specified Mash distance threshold. The Mash distance can be compensated based on plasmid size differences. For prospective surveillance, Early Warning Alerts (EWAs) can be defined, which are automatically triggered when a plasmid of a sample newly processed via the pipeline matches a plasmid already in the database (excluding likely clonal events). These EWAs can be limited by (spatiotemporal) metadata. The results of plasmid searches and EWAs are displayed in Plasmid Tables. The plasmid table contains information on rep-type(s), integrated mobile genetic elements (iMGEs), and AMR targets for each plasmid. Where appropriate after removing some plasmid(s) the plasmid search table can be stored as a plasmid snapshot. Finally, plasmids can be aligned and visually compared using an integrated version of pyGenomeViz. To facilitate downstream analyses, the Fix Start and Orientation for Plasmids or Chromosomes function can be enabled in the pipeline script to automatically re-start and -orient circular contigs.
For a demonstration it is referred to the Tutorial for Real-time Plasmid Transmission Detection and Alert from Long-read Sequencing Data.
Requirements
The Long-read Data Plasmid Transmission Analysis Module is part of the extra charged Long-read Data Analysis Bundle [LDAB].
![]() Important:
Important:
- If SeqSphere+ is installed on Windows, the Long-read Data Plasmid Transmission functionality requires the Windows Subsystem for Linux (WSL).
- If SeqSphere+ is installed on Linux, the Long-read Data Plasmid Transmission functionality must once be installed by calling the installation of Bioinformatic Tools on Linux.