| wiki | search |
Contents |
1 Overview
This tutorial describes how to use the Ridom SeqSphere+ software to analyze next-gen sequencing data with a rapid local 'ad hoc' MLST+ schema.
Neisseria meningitidis is used exemplarily for this demonstration. However, by reading this tutorial you should be able to define your own projects for other species.
This tutorial requires the 64bit version of Ridom SeqSphere+.
2 Preliminaries
- Installation: This tutorial requires a running SeqSphere+ client and server. If not done yet: Download and install the SeqSphere+ 64bit client and server software on your computer. Start the SeqSphere+ server, then start the SeqSphere+ client and initialize the database. For evaluation purpose a free evaluation license can be requested.
- Tutorial Data: Download the example data archive SeqSphere_Examples_PGM_200bp_N_meningitidis.zip for this tutorial, and extract the zip-file on your computer. The example data contains Neisseria meningitidis PGM 200bp whole genome shotgun (WGS) data of 3 samples of a community outbreak that was published by U. Vogel et al. (J. Clin. Microbiol. 50: 1889, 2012).
3 Finding Closely Related Reference Genome
Each MLST+ schema is based on a reference genome. This reference genome must be finished and annotated. For highest discriminatory power choose a closely related reference strain/genome. Therefore the following tutorial uses MLST to find a closely related reference genome.
3.1 Create MLST Task Template
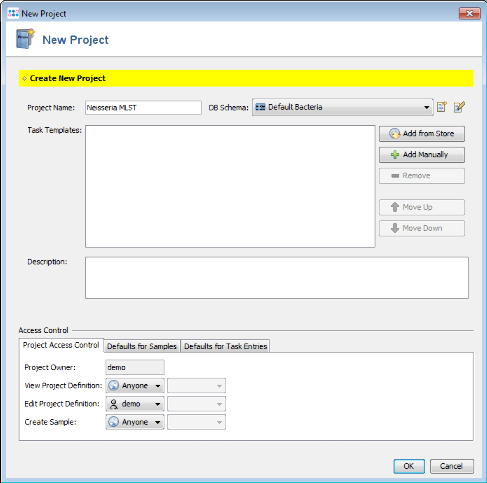
- Step 1: Create a new Project with the menu: File | New | Project
- Step 2: Enter a name for your Project (e.g., Neisseria MLST)
- Step 3: Press
 Add Manually in Task Templates section.
Add Manually in Task Templates section.
- Step 4: Press
 Create New top open the Task Template wizard dialog.
Create New top open the Task Template wizard dialog.
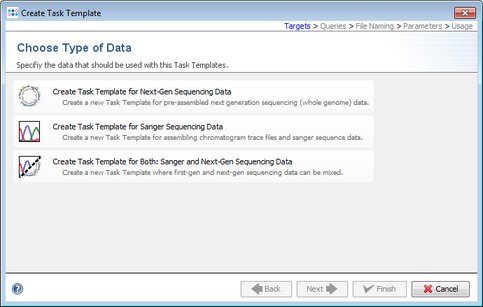
- Step 5: Choose Create Task Template for Next-Gen Sequencing Data.
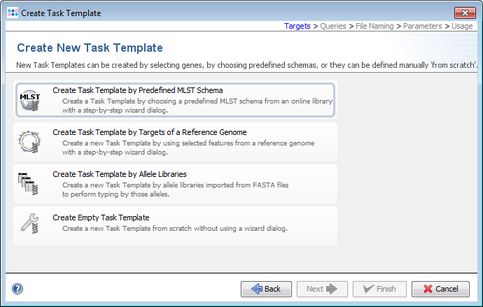
- Step 6: Now choose Create Task Template by Predefined MLST Schema.
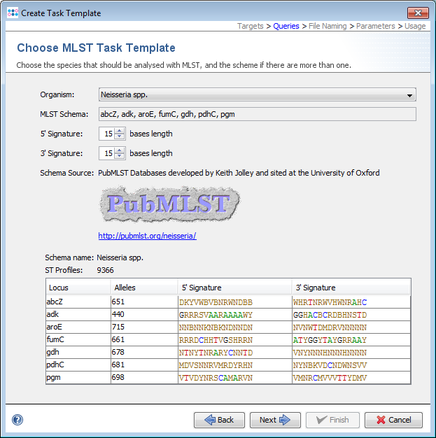
- Step 7: Choose the organism Neisseria spp. in the selection box. The data is downloaded from the public MLST server. Click Next to show the Target Parameters.
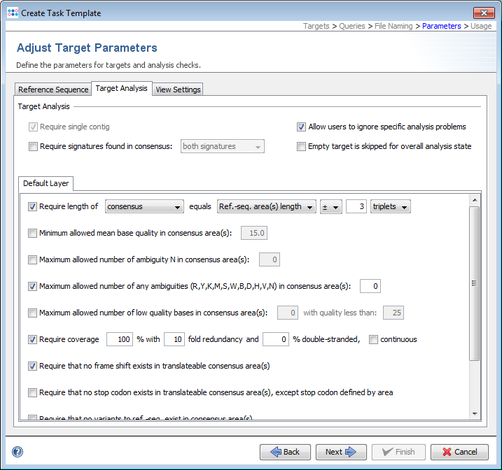
- Step 8: The Target Parameters for the Task Template (e.g., the quality check parameters) can be left unchanged. Click Next to continue.
- Step 9: Check the name of your new Task Template, and confirm with Finish.
- Step 10: Press OK to save the new Task Template and add it to your Project. Finally save your Project by clicking OK in the Project window.
3.2 Add NCBI GenBank Genomes to MLST Project
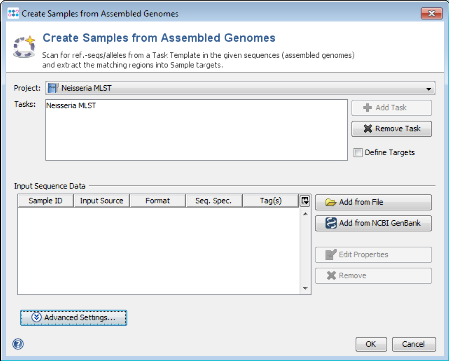
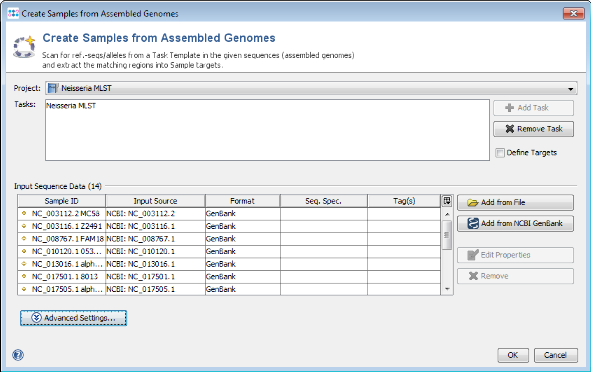
- Step 1: Choose from menu File | Create Samples from Assembled Genomes.
- Step 2: Choose the Project you just created.
- Step 3: Press
 Add from NCBI GenBank
Add from NCBI GenBank
- Step 4: Press
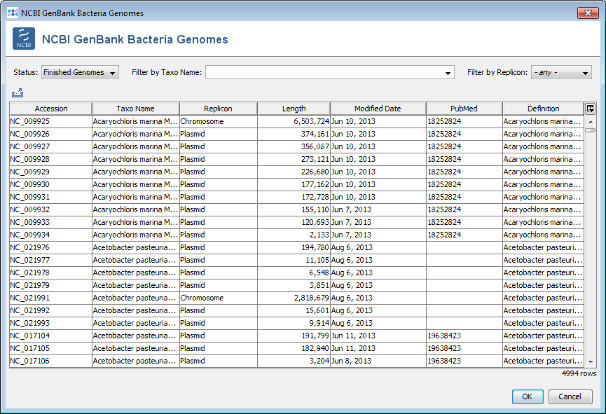
 Search button. A list of all finished genomes that are available on the NCBI GenBank ftp site is shown.
Search button. A list of all finished genomes that are available on the NCBI GenBank ftp site is shown.
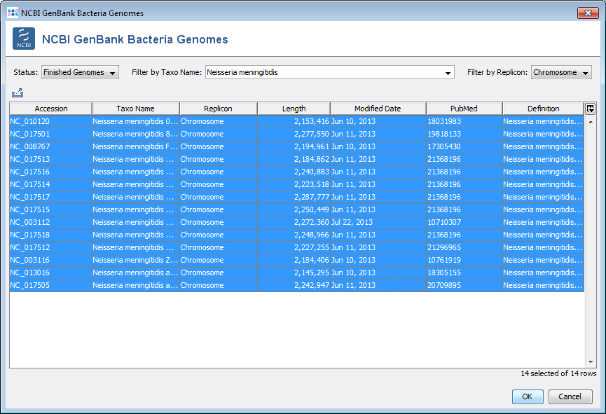
- Step 5: Choose or enter Neisseria meningitidis in the Taxo Name field and select Chromosome in the Replicon field to filter the list. Select all found Neisseria chromosomes and press two times OK to download the entries.
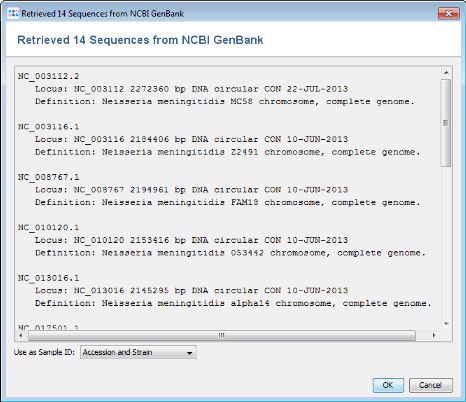
- Step 6: A preview of the NCBI GenBank datasets (14 as of November 2013) that will be downloaded is shown. Press OK to continue.
- Step 7: The NCBI GenBank genomes are added to the import list. (Do not yet press the OK button!)
3.3 Add Outbreak Strains to MLST Project
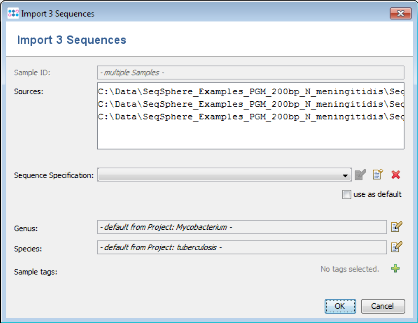
- Step 1: Now use the button
 Add from File and choose the four .ace.gz-files from the tutorial data folder. Those files are de-novo assemblies (assembled with MIRA) of NGS data (PGM 200bp).
Add from File and choose the four .ace.gz-files from the tutorial data folder. Those files are de-novo assemblies (assembled with MIRA) of NGS data (PGM 200bp).
- Step 2: The upcoming window allows to specify sequencing information. Skip this step by clicking OK. Confirm again with OK to start the import process.
- Step 3: The sequence data of the 3 isolates is added to the bottom of the import list
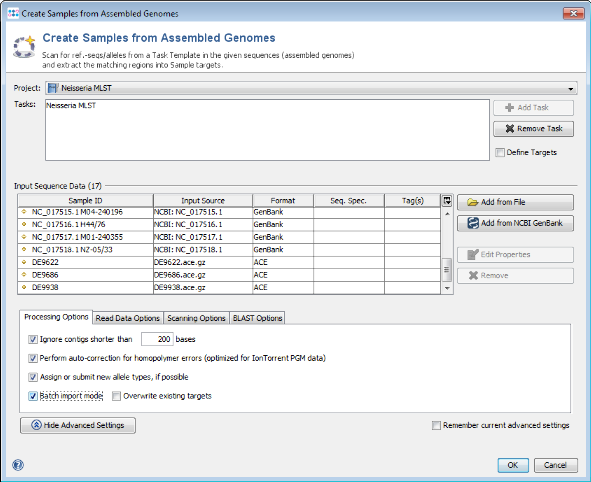
- Step 4: Press the button Advanced Settings and mark the option Batch import mode.
- Step 5: Press OK to start the import.
3.4 Comparing Imported Strains
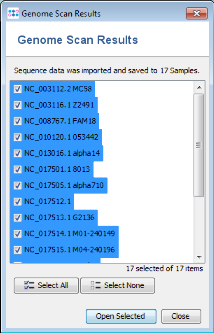
- Step 1: After the import process is finished, a list of the imported Samples is shown. Press just Close here, because it is not necessary to open the Samples for comparing them.
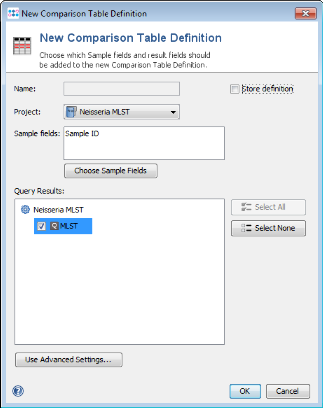
- Step 2: From menu Tools | Comparison Table.
- Step 3: Press New Definition.
- Step 4: Uncheck Store definition, choose the project where the data was just imported in, and mark the check box of MLST in the query results section. Press OK to continue.
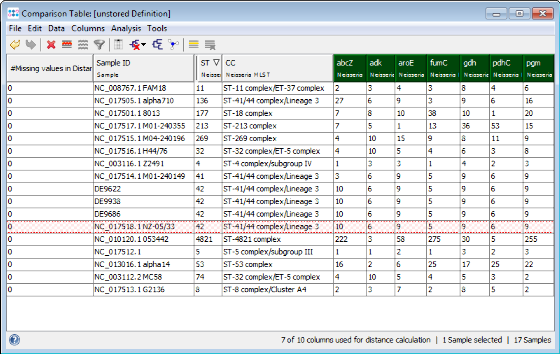
- Step 5: All three outbreaks strains have the MLST ST 41. There also one strain from NCBI GenBank with this ST: NC_017518 NZ-05/33
- Step 6: Press the
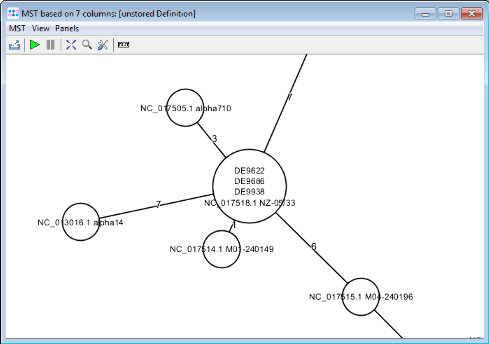
 Minimum Spanning Tree button in the toolbar to calculate the distances between the Samples and draw a minimum spanning tree for them. This tree also shows that NC_017518 NZ-05/33 can be used as closely related reference genome.
Minimum Spanning Tree button in the toolbar to calculate the distances between the Samples and draw a minimum spanning tree for them. This tree also shows that NC_017518 NZ-05/33 can be used as closely related reference genome.
4 Creating Rapid local 'ad-hoc' MLST+ Schema
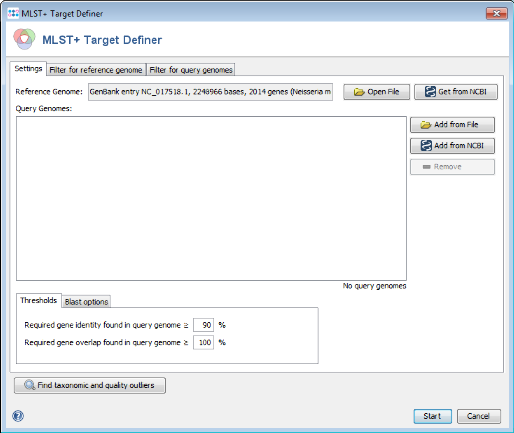
- Step 1: From menu Tools | MLST+ Target Definer.
- Step 2: Press Get from NCBI in the Reference Genome field on the top and the accession number of the found strain: NC_017518. Press OK to retrieve the sequence and insert it as reference genome.
- Step 3: Press the Start button to begin the target finder process.
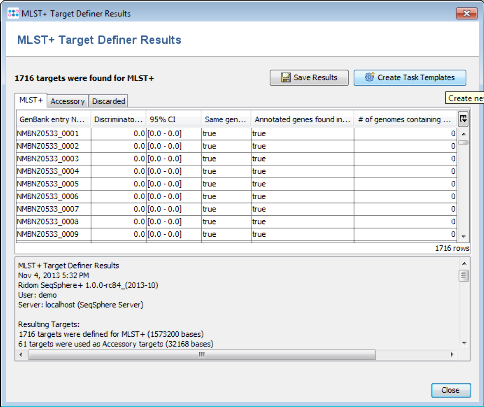
- Step 4: After a few minutes the process is finished and the results are shown. Each of the 2014 genes of NC_017518 was added to one category: MLST+, Accessory or Discarded.
- MLST+ contains 1716 genes that are not homologous, do not contain stop codons and do not overlap with other genes. Those genes are used as targets for MLST+.
- Accessory contains 61 genes that are not homologous and do not contain stop codons, but overlap with other genes. By convention, those genes are not used for MLST+. However, they can be used in addition to increase the discriminatory power if the resolution of MLST+ is not high enough.
- Discarded contains 237 genes that are homologous or contain stop codons. Those genes are not used at all.
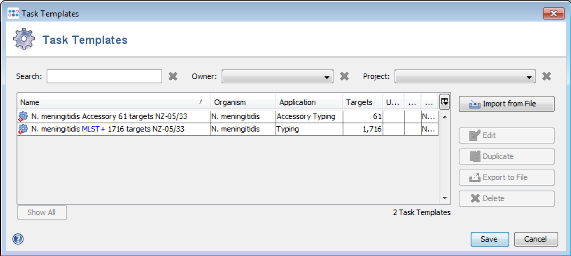
- Step 5: Press the button Create Task Templates to create two Task Templates for the target definer results that can be used for analyzing N. meningitidis next-gen sequencing data: The MLST+ Task Template is created for the 1716 MLST+ targets. A second Task Template called Accessory is created for the 61 genes of the Accessory category.
- Step 6: Press the Save to store the Task Templates.
- Step 7: Press the Yes to create a new Project for the Task Templates. The name of the Project can be changed.
Ridom SeqSphere+ is a resequencing software. Once you have setup your Project with the Task Templates you can literally analyze hundreds/thousands of sequence data automatically.
5 Import Outbreak Strains to MLST+
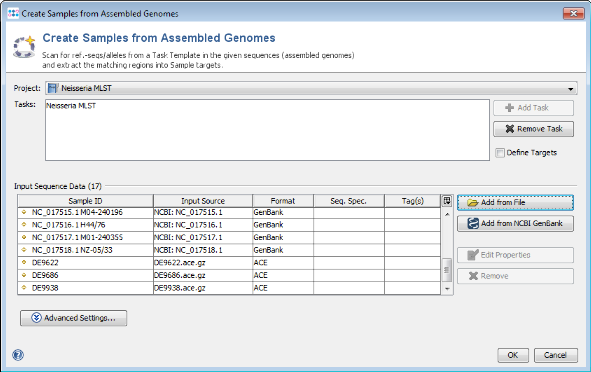
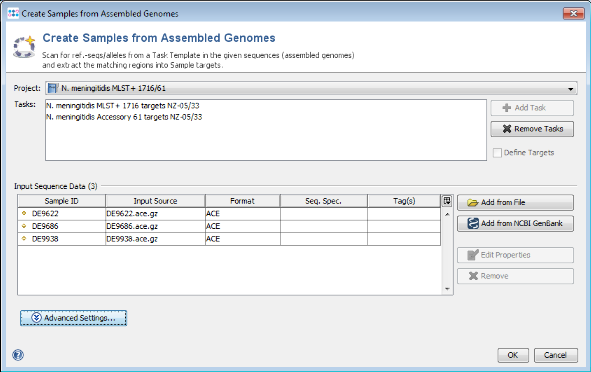
- Step 1: Choose from the menu File | Create Samples from Assembled Genomes
- Step 2: The new MLST+ Project you just created and the two Task Templates should be preselected already.
- Step 3: Now use the button
 Add from File and choose the three .ace.gz-files from the tutorial data folder. Those files are de-novo assemblies (assembled with MIRA) of NGS data. The upcoming window allows to specify sequencing information. Skip this step here by clicking OK. Confirm again with OK to start the import process.
Add from File and choose the three .ace.gz-files from the tutorial data folder. Those files are de-novo assemblies (assembled with MIRA) of NGS data. The upcoming window allows to specify sequencing information. Skip this step here by clicking OK. Confirm again with OK to start the import process.
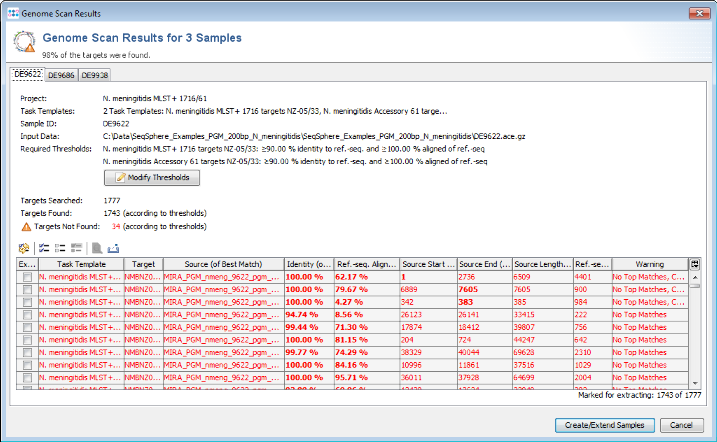
- Step 4: Ridom SeqSphere+ now loads all input sequences and finds (by using built-in BLAST) each of the target reference sequences that are defined in the Task Template. After this step has been done, a preview window with scanning result for each input sequence file is shown (not shown in batch mode). A table per Sample lists all the targets with the BLAST hit results (percent identity, alignment, start and stop positions) and other relevant data. The first column of table marks the targets that should be imported into SeqSphere+. By default, only the targets that fulfill the specified identity and alignment thresholds (e.g., 90% identity to ref.-seq., and 100% aligned of ref.-seq.) will be added to the new Sample entry. The targets that don't have a unique match that fulfills the thresholds are colored red. The thresholds are normally taken from the Task Template, but they also can be changed in this step. This preview can be left to default and just confirmed with the Create/Extend Samples button.
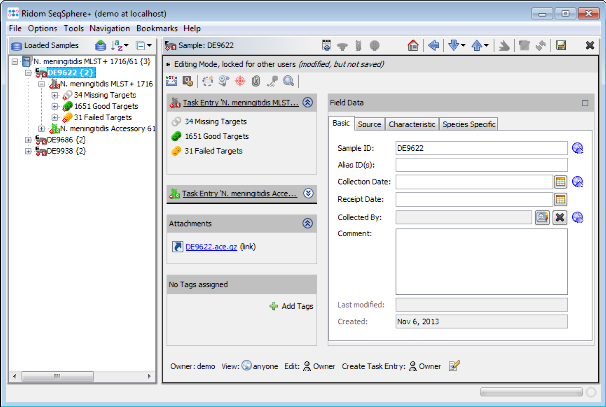
- Step 5: After the import is completed, the navigation tree shows all new Samples. Each Sample node in the navigation has two sub nodes: The MLST+ task and the Accessory task. Below the task nodes there are the target nodes. Each target node represents one sequence (often a gene) extracted from the input data (genomes or wgs contigs). The targets can have different states:
-
 Missing Targets (gray) were not extracted (because the match did not reached the thresholds in the previous step)
Missing Targets (gray) were not extracted (because the match did not reached the thresholds in the previous step)
-
 Good Targets (green) were extracted and fulfill all requirements that are defined in the Task Template Analysis Parameters.
Good Targets (green) were extracted and fulfill all requirements that are defined in the Task Template Analysis Parameters.
-
 Failed Targets (yellow) were extracted, but fail at least in one of the requirements that are defined in those parameters. For example, they may have frame shifts and incorrect lengths compared to the published FAM18 strain sequence. Those targets must be inspected further.
Failed Targets (yellow) were extracted, but fail at least in one of the requirements that are defined in those parameters. For example, they may have frame shifts and incorrect lengths compared to the published FAM18 strain sequence. Those targets must be inspected further.
-
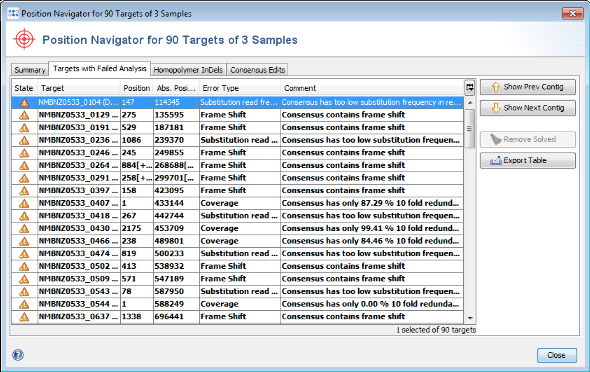
- Step 6: Select Tools | Position Navigator from the menu. Initially all yellow targets are selected, so just click OK. After a short while the Position Navigator is opened. This window shows the most interesting positions in the new sequences.
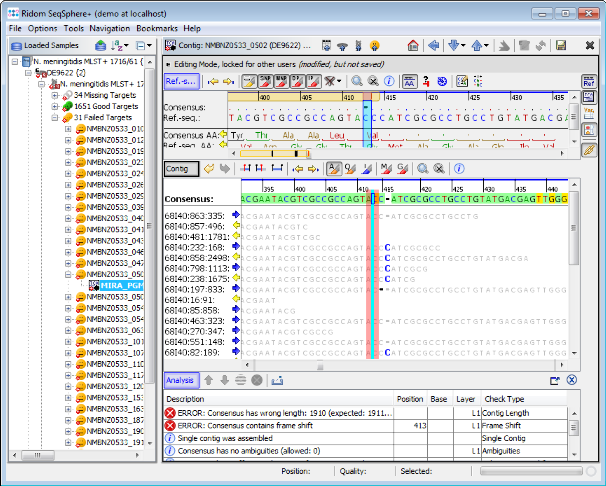
- Step 7: Double click on a row in the table to jump to the according position in the contig. Errors that cannot be corrected automatically must be manually edited in the contig. Changes can be made to the sequence itself here by right-clicking on the sequence and choosing form the options in the context menu, Add base, Delete base, etc. For example, if many stop codons exist in contig, most likely there is a frame shift. This can be found by scrolling through the list of analysis problem locations.
- Step 8: Choose from the menu
 File | Save All to store the 3 Samples to the database on your SeqSphere+ server.
File | Save All to store the 3 Samples to the database on your SeqSphere+ server.
6 Compare Outbreak Strains with MLST+
- Step 1: Choose from the menu Tools | Comparison Table
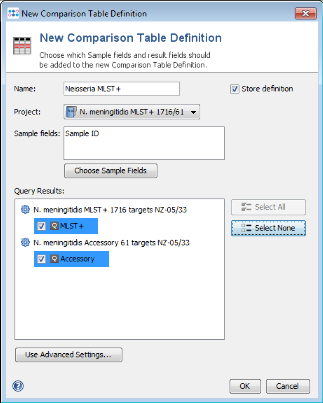
- Step 2: Press New Definition.
- Step 3: Enter a name for the Comparison Table (e.g., Neisseria MLST+), choose the new MLST+ project and press Select All in the query result section to include all imported targets. Press OK and confirm with Save all Changes to store the new Samples in the database.
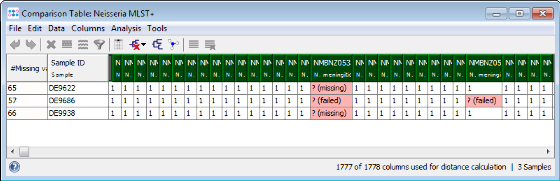
- Step 4: The comparison table is opened and shows the data for the three Samples. The columns with a green header are used for distance calculation. Table cells with red background contain missing values. Those appear if a MLST+ target was not found at all in an input sequence ("? (missing)"), or if the analysis for this target has failed, e.g., because of a frame shift error ("? (failed)").
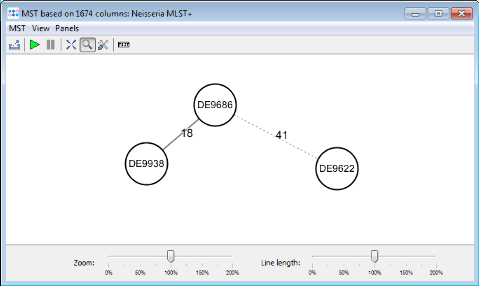
- Step 5: Press the
 Minimum Spanning Tree button in the toolbar to calculate the distances between the Samples and draw a minimum spanning tree for them. If the table contains missing data (targets that have no allele types assigned yet), the columns can be automatically removed from distance calculation by selecting Remove Columns from Distance Calculation.
Minimum Spanning Tree button in the toolbar to calculate the distances between the Samples and draw a minimum spanning tree for them. If the table contains missing data (targets that have no allele types assigned yet), the columns can be automatically removed from distance calculation by selecting Remove Columns from Distance Calculation.
- The final minimum spanning tree shows allele distances between the three outbreak strains which are higher than the ones that can be calculated with the stable MLST+ schema.