Contents
Overview
The Procedure Details are used to document the laboratory procedure, the assembly procedure, the target scan procedure, and the target QC procedure.
The laboratory and assembly procedure details can be predefined and managed using the menu item Options | Procedure Details. If a new Sample is created from genome data, they can be assigned to Samples. The laboratory and assembly procedure details of an existing Sample can also be edited.
The procedure details are also shown in the Procedure tab of the Sample Overview and they can be exported like epidemiological data fields. The laboratory and assembly procedure details can also be exported to and imported from SPEC Files.
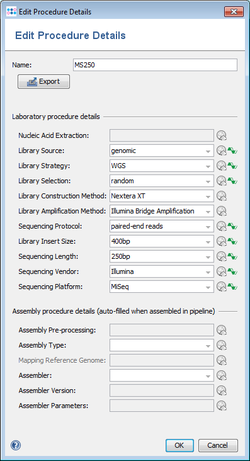
Laboratory Procedure Details
The laboratory procedure details must be specified manually for an input file or defined once in a pipeline script for all following processed samples.
| Field | Description | Submission |
|---|---|---|
| Nucleic acid extraction | Link to a literature reference, electronic resource or a standard operating procedure (SOP) | |
| Library source | Library source used (e.g., genomic, transcriptomic, cDNA, or metagenomic) | |
| Library strategy | Library strategy used (e.g., WGS, WGA, WXS, or RNA-Seq) | |
| Library selection | Library selection used (e.g., random, PCR, random PCR, orRT-PCR) | |
| Library construction method | Library construction method used | |
| Library amplification method | Library amplification method used | |
| Sequencing protocol | Sequencing protocol used (e.g., single-end reads, paired-end reads, or mate-pair reads) | |
| Library insert size | Library insert size in base pairs (excluding adaptors and/or primers) | |
| Sequencing length | Number of bases of insert sequenced | |
| Sequencing vendor | Sequencing platform producer (e.g., Illumina, Ion Torrent, Pacific Biosciences, or 454) | |
| Sequencing platform | Sequencing platform used (e.g., Illumina MiSeq or Ion Torrent PGM) |
Submission Icons:
![]() Potentially this field is submitted and published on cgMLST.org when the Sample is submitted.
Potentially this field is submitted and published on cgMLST.org when the Sample is submitted.
![]() This field is submitted (required) and published on EMBL-EBI ENA when the Sample is submitted.
This field is submitted (required) and published on EMBL-EBI ENA when the Sample is submitted.
Assembly Procedure Details
The laboratory procedure details can be specified manually for an input file or defined once in a pipeline script for all following processed samples. If the pipeline performs an assembling (de novo or mapping), any existing values will be overwritten by the actually used assembly procedure details.
| Field | Description | Submission |
|---|---|---|
| Assembly pre-processing | Coverage downsampeled and/or trimmed by quality (window and QV) | |
| Assembly type | General assembly approach (de-novo or mapping) | |
| Mapping reference genome | NCBI accession number of reference genome against which raw read data were mapped | |
| Assembler | Software used for assembly | |
| Assembler version | Version of software used for assembly | |
| Assembler parameters | Assembler parameters used for assembly | |
| Sequencing comment | Additional information regarding laboratory and assembly meta-data | |
| Downsampled to Coverage1) | Expected genome coverage that was used to calculate the ratio for random downsampling | |
| Expected Genome Size for Downsampling1) | Expected genome size that was used to calculate the ratio for random downsampling | |
| Quality Trimming1) | Parameters that were used for quality trimming of reads |
1) not editable and only available if assembled with a SeqSphere+ pipeline
![]() Potentially this field is submitted and published on cgMLST.org when the Sample is submitted.
Potentially this field is submitted and published on cgMLST.org when the Sample is submitted.
Target Scan Procedure Details
The target scan procedure details are stored as one text field. In contrast to the laboratory and assembly details, these details are not editable and never submitted to cgMLST.org. They are always automatically filled in by SeqSphere+. They are defined in the Task Procedure settings of the Task Template.
Example:
Ridom SeqSphere+ version 3.4.0_(2016-10)
BLAST version 2.2.12
Ignore contigs shorter than 200 bases
Scan for ref.-seqs. of L. monocytogenes cgMLST v2.1
ref.-seq. scan BLASTN options: word size=11, mismatch=-1, match=1, gap open=5, gap ext.=2
scan thresholds for hit: identity>=90, aligned=100
require unique match found within the scan thresholds
Scan for ref.-seqs. of L. monocytogenes Accessory v2.0
ref.-seq. scan BLASTN options: word size=11, mismatch=-1, match=1, gap open=5, gap ext.=2
scan thresholds for hit: identity>=90, aligned=100
require unique match found within the scan thresholds
Scan for ref.-seqs. and alleles of L. monocytogenes MLST v1.0
ref.-seq. scan BLASTN options: word size=11, mismatch=-1, match=1, gap open=5, gap ext.=2
allele scan BLASTN options: word size=11, mismatch=-1, match=1, gap open=5, gap ext.=2
scan thresholds for hit: identity>=90, aligned>=99
require unique match found within the scan thresholds
Target QC Procedure Details
The target QC procedure details are stored as one text field. In contrast to the laboratory and assembly details, these details are not editable and never submitted to cgMLST.org. They are always automatically filled in by SeqSphere+. They are defined in the Target QC Procedure settings of the Task Template.
Example:
Ridom SeqSphere+ version 3.4.0_(2016-10)
Target QC Procedure for L. monocytogenes cgMLST v2.1
require that length of consensus equals ref.-seq. area(s) length +-3 triplets
require that no ambiguities (R,Y,K,M,S,W,B,D,H,V,N) are in the consensus area(s)
require that no frame shift exists in the translatable consensus area(s)
Target QC Procedure for L. monocytogenes Accessory v2.0
require that length of consensus equals ref.-seq. area(s) length +-3 triplets
require that no ambiguities (R,Y,K,M,S,W,B,D,H,V,N) are in the consensus area(s)
require that no frame shift exists in the translatable consensus area(s)
Target QC Procedure for L. monocytogenes MLST v1.0
require that length of consensus equals ref.-seq. area(s) length +-3 triplets
require that no ambiguities (R,Y,K,M,S,W,B,D,H,V,N) are in the consensus area(s)
require that no frame shift exists in the translatable consensus area(s)