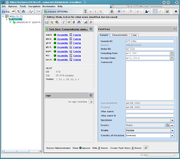
The Sample Overview shows a summary of all data stored within the Sample entry.
The view is organized in two columns. The left columns contains
Task Entry Summaries
In the left column the summaries for all Task Entries of the Sample are listed. Click on the drop down icon to expand the view and show the details for the targets and results of Task Entry
Sample Tags
Below the tags are shown that are assigned to the Sample. Use the button to add or remove tags.
Sample Data
The right column shows a tabbed pane with multiple tabs.
- The Epi tabs show the fields defined by the Database Scheme of the Project. With the default schemes those tab are Epi Basic, Epi Source, and Epi Characteristic, and for some species the Epi Species Specific. If condition checks were defined in the Database Scheme, there might be errors and warnings. They are shown in a text field at the bottom of the editor and as icons in the header of the tabs:
- Errors are shown by a red border of a data field and an error sign
 in the tab.
in the tab.
- Warnings are shown by a yellow border of a data field and a warning sign
 in the tab.
in the tab.
- Errors are shown by a red border of a data field and an error sign
- The Procedure tab shows the proccedure details, the procedure statistics and the procedure files of this Sample orderd by the procedure that were performed.
- The
 Results tab shows a summary of important result fields of all Task Entries of the Sample and a button to create Sample reports. Created and stored Sample reports are listed at the top of the Results panel.
Results tab shows a summary of important result fields of all Task Entries of the Sample and a button to create Sample reports. Created and stored Sample reports are listed at the top of the Results panel.
Toolbar
The toolbar on the top of the panel shows the following buttons:
-
 Export consensi: Export the consensus sequences of all Task Entries of this Sample into a FASTA library file.
Export consensi: Export the consensus sequences of all Task Entries of this Sample into a FASTA library file.
-
 Reassemble Reads: Re-Assemble the read sequences (e.g., chromatograms) of all Task Entries of this Sample. This feature is only available for Sanger sequencing data.
Reassemble Reads: Re-Assemble the read sequences (e.g., chromatograms) of all Task Entries of this Sample. This feature is only available for Sanger sequencing data.
-
 Restart Queries: Restart the queries of all Task Entries of this Sample.
Restart Queries: Restart the queries of all Task Entries of this Sample.
-
 Add Sequences from Assembly File: Open the Process Assembled Genome Data window with the WGS data file linked or attached to the Sample. This feature is only available for whole genome sequencing data.
Add Sequences from Assembly File: Open the Process Assembled Genome Data window with the WGS data file linked or attached to the Sample. This feature is only available for whole genome sequencing data.
-
 Show Sample audit trail: Show the Sample audit trail.
Show Sample audit trail: Show the Sample audit trail.
-
 Position Navigator: Open the Position Navigator for all Task Entries of this Sample.
Position Navigator: Open the Position Navigator for all Task Entries of this Sample.
-
 Add attachment: Add an attachment file to this Sample (any kind of file, like pdf).
Add attachment: Add an attachment file to this Sample (any kind of file, like pdf).
-
 Delete Sample: Delete the Sample permanently from the database.
Delete Sample: Delete the Sample permanently from the database.
-
 Search Samples that have similar allele type combinations in database: Search for similar Samples. Note that the search is based on number of similar allele types, not on sequence similarity.
Search Samples that have similar allele type combinations in database: Search for similar Samples. Note that the search is based on number of similar allele types, not on sequence similarity.
Access Control
On the bottom, specific access rights can be set for the Sample.