General
Allele Typing queries are essentially the same technique as MLST, but are based on whole genome sequence data, and often use a number of targets much larger (e.g. 3000 targets) than basic MLST (using only 7 targets).
This kind of query libraries can additionally have a forth query result state:
-
 new alleles can be assigned either using the Assign Alleles button in the result for or using the menu action
new alleles can be assigned either using the Assign Alleles button in the result for or using the menu action  File | Assign or Submit new Alleles.
File | Assign or Submit new Alleles.
Definition Panel
The definition panel is used to define and edit the query. The following fields can be set in the upper part of the panel:
- Query Name
- A name for this query. This will also be used as category name for the result fields that are filled by this query.
- Execution Mode
- Perform automatically: Every time the typing is changed, all opened Samples are re-typed.
- Never perform automatically: Re-typing has to be started manually using a Sample's context menu.
- Comment
- A comment for this query. Only shown in the definition panel.
The allele libraries are not managed in the Task Template editor. The command Manage Allele Libraries can be used to manage the libraries.
Result Panel
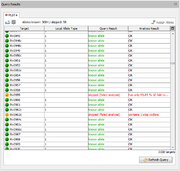
The result panel that is a part of the Task Entry overview shows a table where each row represents a target. The columns are:
- Target
- The target
- Source
- The source of the sequences, for next-gen data this is the whole-genome-sequencer contig from which the sequence was extracted.
- Allele Type
- The matching allele type
- Query Result
- Can have one of the four values: known allele (if the allele was found in library), new allele (if the allele was not found), not done (if the analysis has failed), or no sequence (if no sequence data exists for this target).
- Analysis Result
- if the analysis has failed this shows the first error that was reported from analysis, otherwise it shows 'OK'.
The toolbar above the table shows the following buttons:
-
 Export the table to a file.
Export the table to a file.
-
 Navigate to the contig of the selected target.
Navigate to the contig of the selected target.
-
 Search Samples Locally: Search Samples that use the current Task Template and that have similar allele type combinations in database. Note: the actual DNA sequence of the allele types is not used for similarity calculation. Only the ratio of same allele types and all allele types is used.
Search Samples Locally: Search Samples that use the current Task Template and that have similar allele type combinations in database. Note: the actual DNA sequence of the allele types is not used for similarity calculation. Only the ratio of same allele types and all allele types is used.
-
 Assign Alleles: Add new the allele types to the query libraries.
Assign Alleles: Add new the allele types to the query libraries.