Introduction
The S. aureus spa-typing task template can be used for WGS and for Sanger sequencing data to assign a type based on the Staphylococcus protein A gene (spa). The task template can be downloaded from the Task Template Sphere.
If WGS data is used as input, primer sequences are used to find the spa region in the genome scanning step that is performed before the spa-typing starts (forward primer: TAAAGACGATCCTTCGGTGAGC, reverse primer: CAGCAGTAGTGCCGTTTGCTT).
The spa-typing searches for the 5' and 3' signatures (RCAMCAAAA, TAYATGTCGT) and trims the sequence to them. Then the sequence is searched for known repeats, and for potentially new repeats that are matching a specific pattern. The result is QC controlled by determining a reliablity rate for the spa-typing (see table below).
To guarantee a standardized nomenclature, the spa-types and repeats are named by a global nomenclature that is controlled by the Ridom SpaServer. They are named with an ID which is either a leading "t" (spa-type) or "r" (repeat) followed by a unique number.
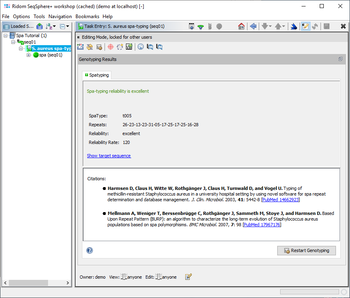
The Task Entry Overview for a spa-typing task shows a result message colored by the reliability, the four result fields, and potentially links to unreliable positions in the sequence that need to be checked.
If an unknown spa-type is found for Sanger sequencing data, and the reliability is "good" or "excellent" (reliability rate >= 100), the new spa-type can be submitted to the SpaServer by using the submission button in the Task Entry overview. New spa-types that were found in WGS data cannot be submitted.
The following meta-data fields are submitted together with the sequence data and the spa-typing result:
- Submitter User ID (database ID and login name) *
- Submitter Email Address *
- Submitter Organisation *
- Submitter City *
- Submitter Country *
- Sample ID *
- Collection Date *
- Country of Isolation *
- Origin (Source Type and Source Subtype)
- MRSA/MSSA
* mandatory field
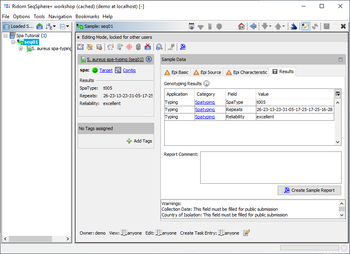
Result Fields
The S. aureus spa-typing provides four result fields:
- SpaType (shown in result table and used in comparison tables for distance calculation)
- Repeats (shown in result table)
- Reliabilty (shown in result table)
- Reliabilty Rate
When the SpaType field is used in a Comparison Table, distances for this field are calculated using the BURP (Based Upon Repeat Patterns) alignment algorithm. A detailed description of the algorithm can be found in Mellmann A, Weniger T, Berssenbrügge C, Rothgänger J, Sammeth M, Stoye J, and Harmsen D. Based Upon Repeat Pattern (BURP): an algorithm to characterize the long-term evolution of Staphylococcus aureus populations based on spa polymorphisms. BMC Microbiol. 2007, 7: 98 [PubMed 17967176], and in Sammeth M, and Stoye J. Comparing tandem repeats with duplications and excisions of variable degree. IEEE/ACM Trans Comput Biol Bioinform. , 3: 395-407 [PubMed 17085848]
Reliability Rating
The reliability of a spa-typing result is specified by a numeric value between 0 and 120. Submitting a strain for a new spa-type requires a reliability of 100 or better. Therefore, new spa-types are not submittable with WGS data.
| Reliability Rating | Input Data | Signature Search | Repeat Search | |
|---|---|---|---|---|
| 120 | excellent | as below | as below | as below, but less than 5 editing steps in repeats |
| 110 | good | consensus of two Sanger sequencing chromatograms files | as below | as below, but no low-quality bases (consensus base quality < 20) |
| 100 | good | at least one Sanger sequencing chromatogram file | as below | as below, but no editing steps in new repeats |
| 90 | sufficient | at least one Sanger sequencing chromatogram file | as below | as below |
| 60 | sufficient | consensus of two Sanger sequencing FASTA files | as below | as below |
| 50 | sufficient | as below | both signatures found on correct positions | as below |
| 40 | sufficient | as below | both signatures found, but position shifted less than 20nt from expected position | as below, but no high quality mismatches (chromatogram base quality≥15) |
| 30 | poor (not reliable) | as below | both signatures found, but position shifted | as below |
| 20 | poor (not reliable) | as below | at least one signature not found, but sequence to short | as below |
| 10 | poor (not reliable) | as below | at least one signature not found | as below, but not more than 5 low-quality positions (consensus base quality < 20) |
| 5 | poor (not reliable) | as below | n/a | continuous repeat succession without gaps |
| 0 | poor (not analyzable) | any FASTA sequence | n/a | no repeats or noncontinuous |