Overview
The proprietary ONT-cgMLST-Polisher is part of the Ridom Typer version 10.5 ONT Data Assembly module. First, it maps the with Dorado SUP basecalled FASTQ reads (>= m4.2 model) to the from Medaka derived assembly consensus FASTA sequence by using minimap2. Next, it scans the alignment for positions in the core and accessory genome MLST genes that might be indicative for methylation related sequencing errors, e.g., differing strand-specific majority consensus calls. Those ‘ambiguous’ positions are then compared against a sequence with a closely related cgMLST allelic profile. Finally, based on the comparison the consensus sequence of ambiguous positions is either confirmed or masked with a ‘N’ call. Thereby, core genome genes that contain a ‘N’ are regarded as missing targets as they fail the quality control that allows by default for no ambiguous bases.
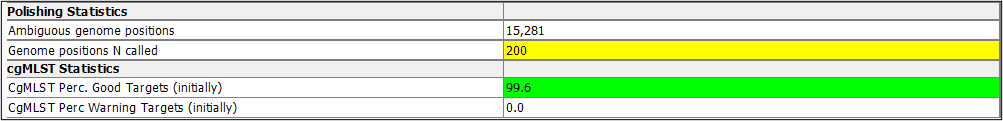
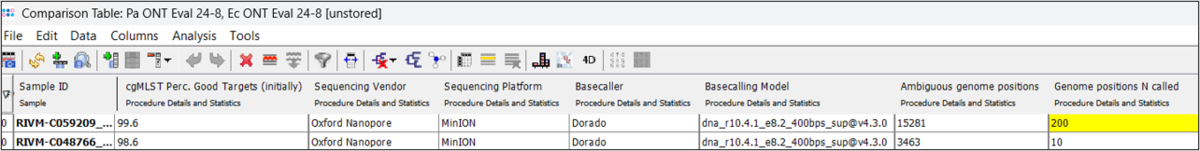
When the ONT-cgMLST-Polisher is used with Dorado SUP m4.2 or m4.3 data, samples with a significantly increased number of called ‘Ns’ are highlighted in yellow to indicate that they may have unreliable results. Data basecalled with Dorado SUP m5.0 or greater are not highlighted.
N.B.: it is not advised to compare closely related samples where only some samples were treated with the ONT-cgMLST-Polisher as this will result in very erroneous trees.
- Significantly increased number of called 'Ns' highlighted in the Polishing Statistics section of the Procedure tab.
- Significantly increased number of called 'Ns' highlighted in the Comparison Table.
ONT-cgMLST-Polisher Accuracies and Contiguities Evaluation and Ring-Trial
For further details, please see ONT-cgMLST-Polisher Accuracies and Contiguities evaluations. In addition, the ONT-cgMLST-Polisher was tested (including with Dorado model 5.0) in a recent ring-trial involving six different laboratories (Prior et al. (2025)).
FOR RESEARCH USE ONLY. NOT FOR USE IN CLINICAL DIAGNOSTIC PROCEDURES.