Contents
Overview
This tutorial describes how to use the Ridom SeqSphere+ software to analyze plasmids with short- and long-read data using the MOB-suite tool.
Preliminaries
- Installation of SeqSphere+: If SeqSphere+ is not available yet, a one-month trial version can be requested. The SeqSphere+ Client and Server software can be installed on the same computer for this tutorial. If you are completely new to SeqSphere+, we recommend to start first with the Tutorial for SeqSphere+ Assembly and cgMLST Analysis Pipeline.
- System Requirements: This tutorial requires at least 8 GB RAM and Windows with an installed Windows Subsystem for Linux (WSL) or a Linux system.
- Tutorial Data: Download the example data archive SeqSphere_Examples_Plasmid_Tutorial.zip (~10 MB) for this tutorial and extract the zip-file on your computer. This zip-file contains FASTA data for the two strains published by van Almsick et al. (Microorganisms 10: 2022) produced with three different sequencing technologies: Illumina (Illumina DNA Prep, MiSeq 2x250bp; de novo assembled with SKESA), Pacific Biosciences (PacBio; HiFi v.3; de novo assembly and following data processing with SMRTLink v. 11), and Oxford Nanopore Technologies (ONT; R.10.4.1 run in accurate mode (260 bps), thresholds for Q Score (>=10) and length (>=200 bp) were set in MinKNOW; de novo assembly with Flye and following Medaka polishing). Illumina and PacBio data were produced at the Univ. Münster and the ONT data by Gabriel Wagner-Lichtenegger from the Univ. Graz, Austria. To demonstrate the import of PacBio run details some artificial run info files were added to the PacBio example data folder. The manuscript describes a presumable intra-host horizontal transfer of a plasmid containing blaCTX-M-27 from a Klebsiella spp. to an Escherichia coli strain with an amplification of the blaCTX-M-27 containing antimicrobial gene cassette.
Define Projects
- Step 1: Start SeqSphere and log in with your account data.
- Step 2: For this tutorial two Projects must be created. Invoke in the menu Options | Projects.
- Step 3: In the upcoming Projects window, press the
 Create new Project icon in the toolbar to start defining a new project.
Create new Project icon in the toolbar to start defining a new project.
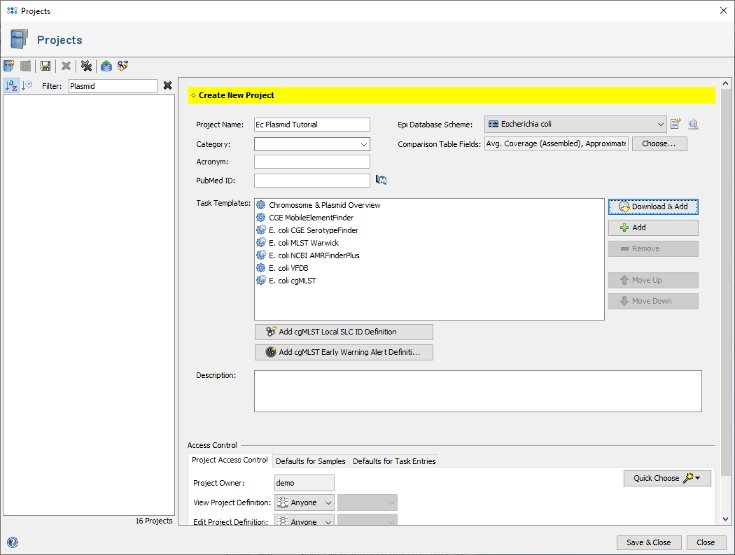
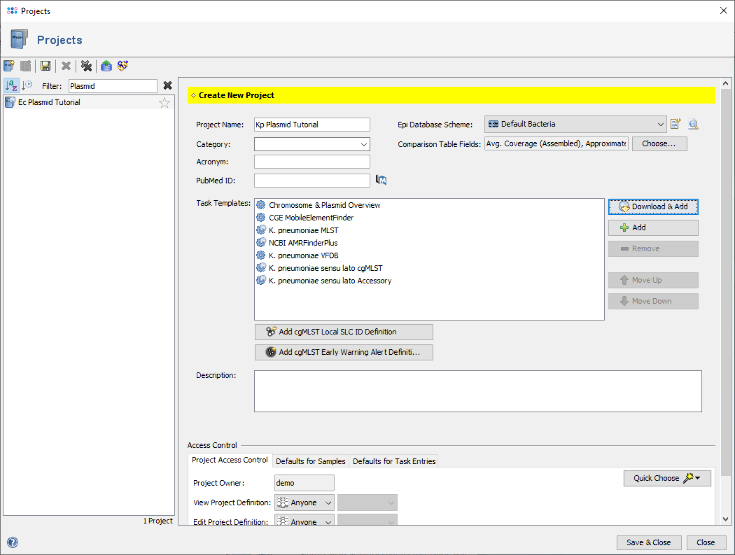
- Step 4: Enter a name for the new Project, e.g., Ec Plasmid Tutorial. Then press
 Download & Add in the Task Templates section to browse the Task Template Sphere.
Download & Add in the Task Templates section to browse the Task Template Sphere.
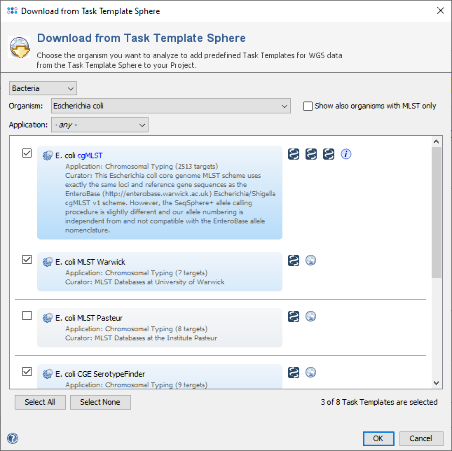
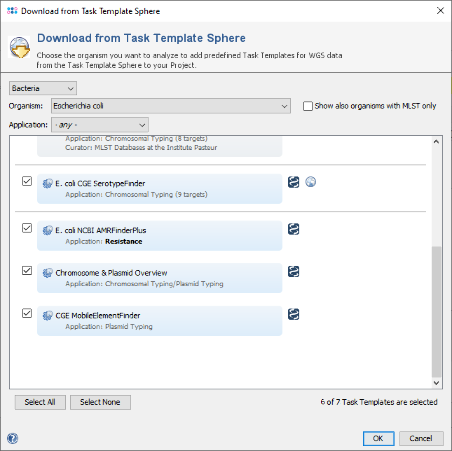
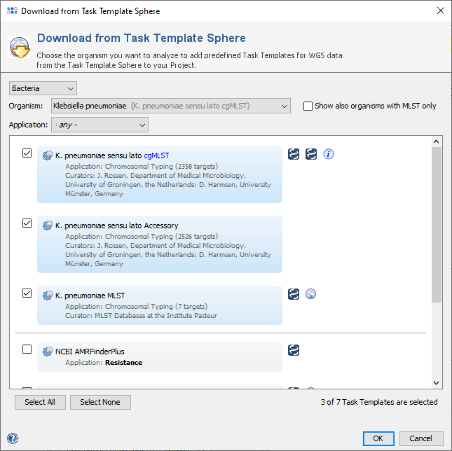
- Step 5: The Task Template Sphere provides all predefined public Task Templates. Choose as organism Escherichia coli. There are eight Task Templates listed, three of them are preselected. Scroll to the bottom of the list, and select also the four Task Templates NCBI AMRFinderPlus, VFDB, Chromosome & Plasmids Overview, and CGE MobileElementFinder by marking the checkboxes left of them. The VFDB task template is used to find virulence factor alleles, the NCBI AMRFinderPlus task template is used to detect antimicrobial resistance-specific genes. The Chromosome & Plasmids Overview and the CGE MobileElementFinder task template are used to reconstruct and characterize plasmids and mobile genetic elements, respectively. The either chromosomal or plasmid location of virulence factors and antimicrobial resistance genes are in addition shown in the Chromosome & Plasmids Overview Task Template. Press OK to confirm the dialog with the seven selected Task templates.
- Step 6: The seven Task Templates are now added to the new project. Press the
 Save icon to store the Project.
Save icon to store the Project.
- Step 7: Now press again the
 Create new Project icon in the toolbar to define a second project. Enter a name again, e.g., Kp Plasmid Tutorial. Then press again
Create new Project icon in the toolbar to define a second project. Enter a name again, e.g., Kp Plasmid Tutorial. Then press again  Download & Add in the Task Templates section.
Download & Add in the Task Templates section.
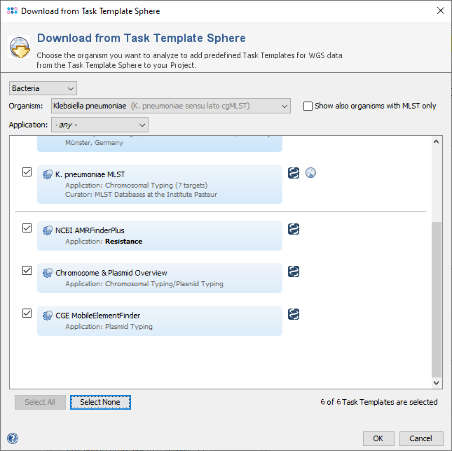
- Step 6: Choose as organism now Klebsiella pneumoniae. There are seven Task Templates listed, three of them are preselected. This time you can just press the Select All button at the bottom of the window to choose all seven Task Templates. Press OK to confirm the dialog and add the Task Templates to your Project.
- Step 7: Finally press the button Save & Close to store also the second Project and to close the dialog.
- Step 8: The Projects are defined now. Continue with the pipeline by invoking in the menu File | Logout & Start Pipeline Mode.
Define and Run Pipeline Script
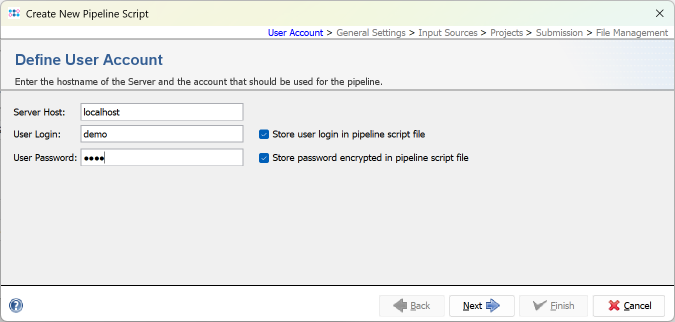
- Step 1: Press Create New Script to open a dialog for creating a new pipeline script. In the first step the Server Host and the User Login must be defined. Just use localhost for your local computer and the same SeqSphere+ user account that you are normally using for the SeqSphere+ login. The option to store user login in the pipeline script is enabled by default. Below enter the User Password of this user account. If wanted, the password can also be stored (encrypted) in the pipeline script. Press Next to move on.
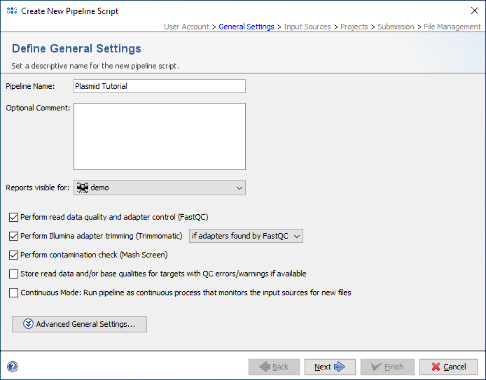
- Step 2: In the Define General Settings panel enter a Pipeline Name (e.g., 'Plasmid Tutorial'). Leave everything else to default and press Next to move on.
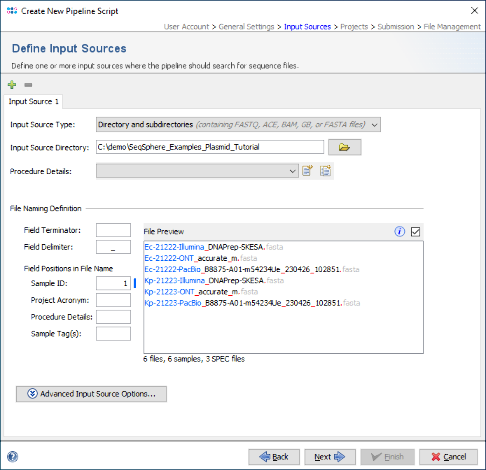
- Step 3: In the next panel the Input Sources for the WGS sequence data are selected. The Input Source Type is preset to Directory. Change this to Directory and subdirectories. Then press the
 button in the directory field below, and select the directory SeqSphere_Examples_Plasmid_Tutorial that was unpacked from the downloaded tutorial data file (see Preliminaries). Enter as Field Delimiter the underscore character "_" to shorten the Sample IDs. The File Preview on the lower right should show the six FASTA files with the Sample IDs highlighted in blue. The Procedure Details can be left empty for this tutorial. The Sequencing Vendor for the example data is set automatically by predefined procedure details files that come with the example data.
button in the directory field below, and select the directory SeqSphere_Examples_Plasmid_Tutorial that was unpacked from the downloaded tutorial data file (see Preliminaries). Enter as Field Delimiter the underscore character "_" to shorten the Sample IDs. The File Preview on the lower right should show the six FASTA files with the Sample IDs highlighted in blue. The Procedure Details can be left empty for this tutorial. The Sequencing Vendor for the example data is set automatically by predefined procedure details files that come with the example data.
- Press Next to move on.
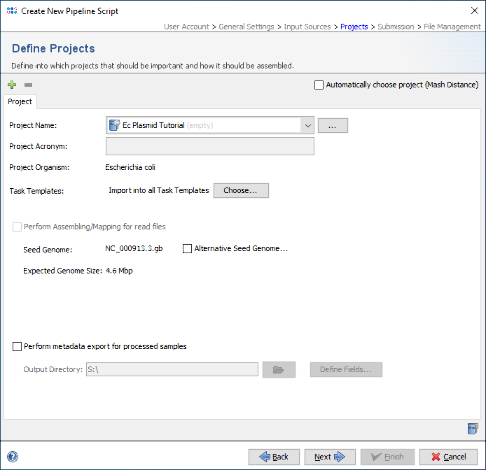
- Step 4: In the Define Projects panel choose the first newly created E. coli Project, i.e., Ec Plasmid Tutorial.
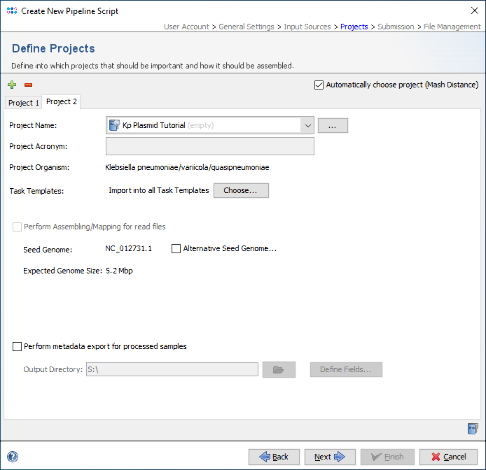
- Step 5: As two different species should processed by this pipeline, select the checkbox Automatically choose project (Mash Distance) at the top of the panel, to detect the Project for each FASTA file automatically by a Mash search against a reference database. Then press the
 Add Project button and choose the second created Project, i.e., Kp Plasmid Tutorial.
Add Project button and choose the second created Project, i.e., Kp Plasmid Tutorial.
- Press Next to move on.
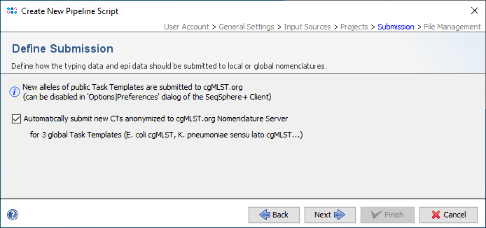
- Step 6: In the Define Submission panel the checkbox to submit new CTs to the public cgMLST Nomenclature Server (cgMLST.org) is already preselected.
- Press Next to move on.
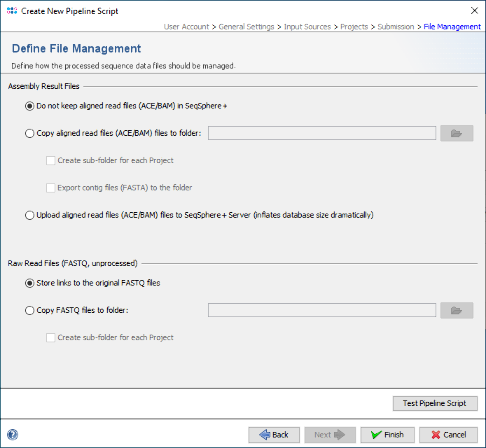
- Step 7: Finally, in the Define File Management panel leave all to default and press Finish to store the new pipeline script.
- Step 8: Now press the button
 Start Script to run the pipeline.
Start Script to run the pipeline.
Analyze Results
- Step 1: After the pipeline is finished restart SeqSphere in interactive mode and log in.
- Step 2: The welcome panel shows a notfication for the finished pipeline. Click in the right part of this notification on the
 dropdown icon, and choose in the upcoming menu the option Load All 6 Samples.
dropdown icon, and choose in the upcoming menu the option Load All 6 Samples.
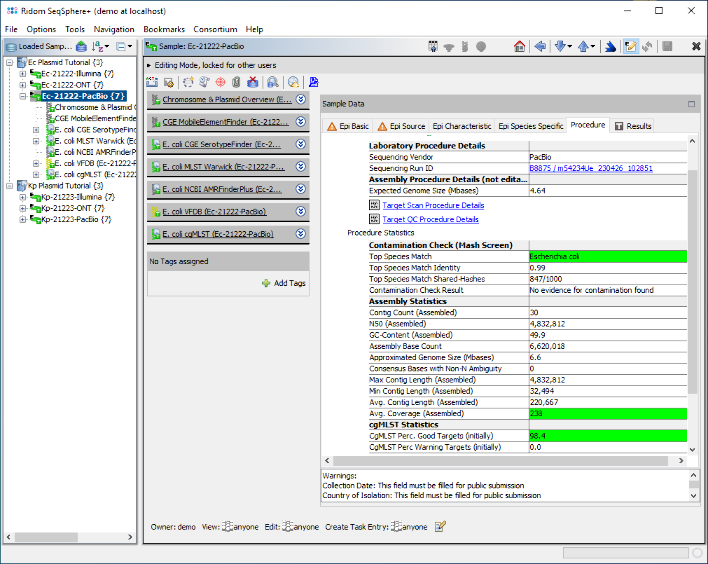
- Step 3: The six Samples are now in the navigation tree. Double-click on the third Sample, the E. coli PacBio Sample Ec-21222-PacBio, to open it in the right panel. Then click on the tab Procedure to see the procedure details for this sample.
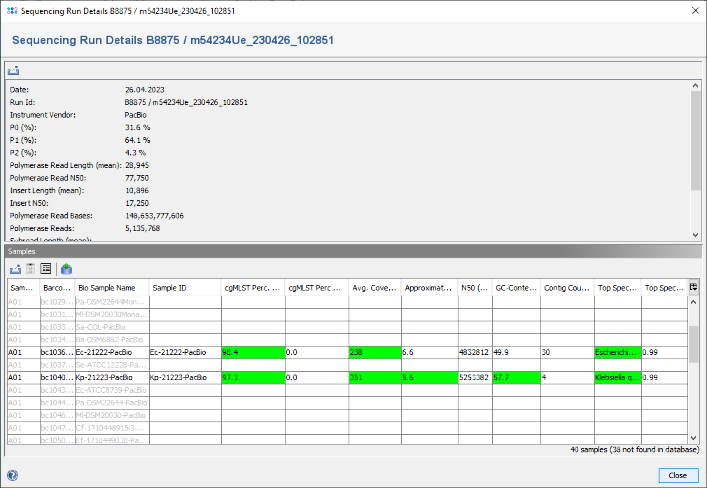
- Step 4: The Procedure Details table contains a link in the line Sequencing Run ID. Click on this link. The PacBio run details are shown that were imported automatically from the accompanying files in the PacBio folder. In the table below it can be seen that only two PacBio samples were imported from this run. The QC related fields are highlighted in green, there are no warnings.
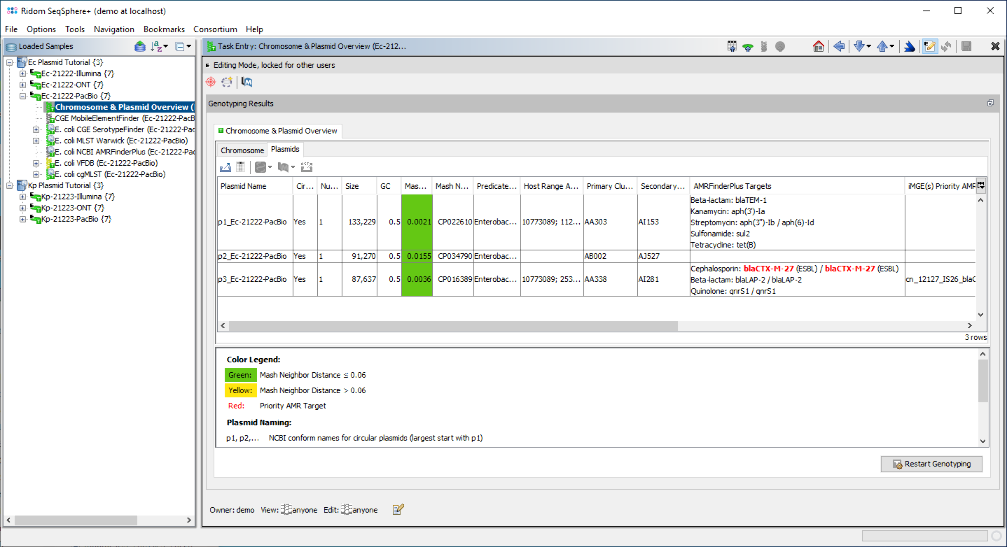
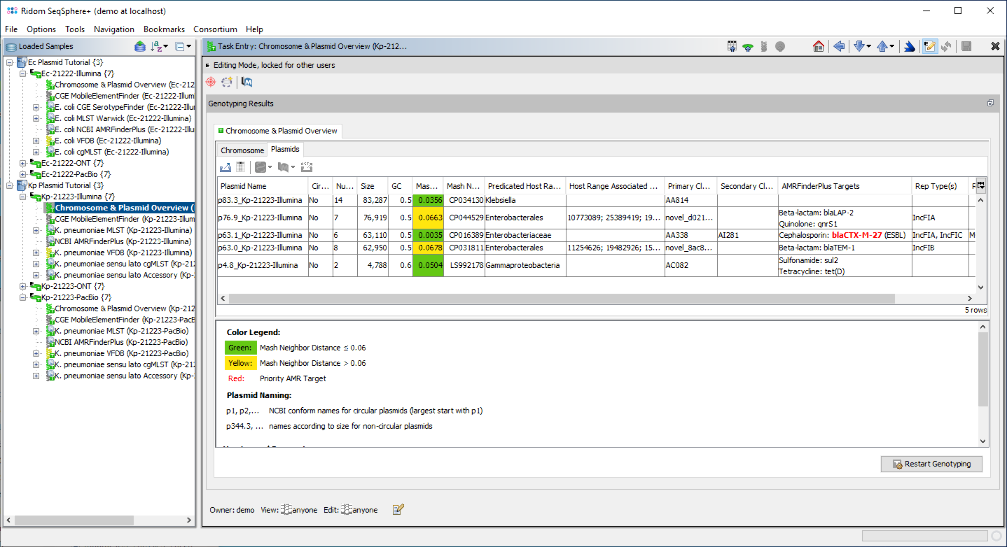
- Step 5: Now close the run info window, and open the Chromosome & Plasmid Overview Task of this Sample. The table shows plasmids that were found/reconstructed by MOB-suite; hereby each row represents a single plasmid and in this case all three plasmids were circular. The column AMRFinderPlus Targets shows that blaCTX-M-27 was found multiple times by AMRFinderPlus in the third plasmid. blaCTX-M-27 is highlighted in red because it is categorized as a priority target. MOB-suite assigned for this 87.6 kb sized plasmid the Secondary Cluster ID AI281. Finally, MobileElementFinder found that this AMR target is part of two nested integrative mobile genetic elements (iMGEs): ISSbo1 and IS26.
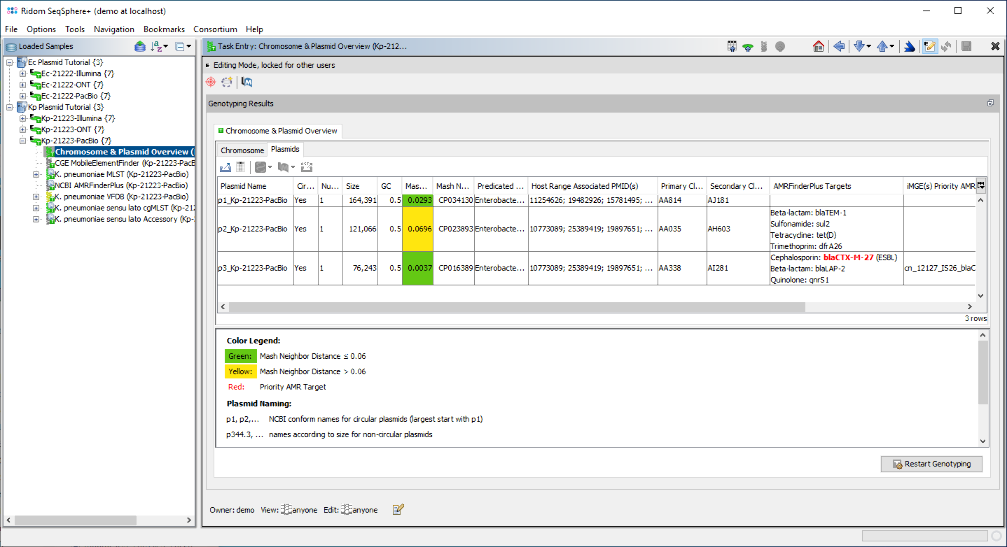
- Step 6: Now open the Chromosome & Plasmid Overview for the last Sample in the list, i.e., the K. pneumoniae PacBio Sample Kp-21223-PacBio. The table shows that a single copy of blaCTX-M-27 was found again on the third circular plasmid that is about 11.4 kb shorter than its E. coli pendant. The Secondary Cluster ID is the same as the one of the E. coli plasmid: AI281.
ONT and Illumina Data
- For the two Samples sequenced with ONT the blaCTX-M-27 targets were also found on plasmids, and the Secondary Cluster ID of these plasmids were correctly AI281.
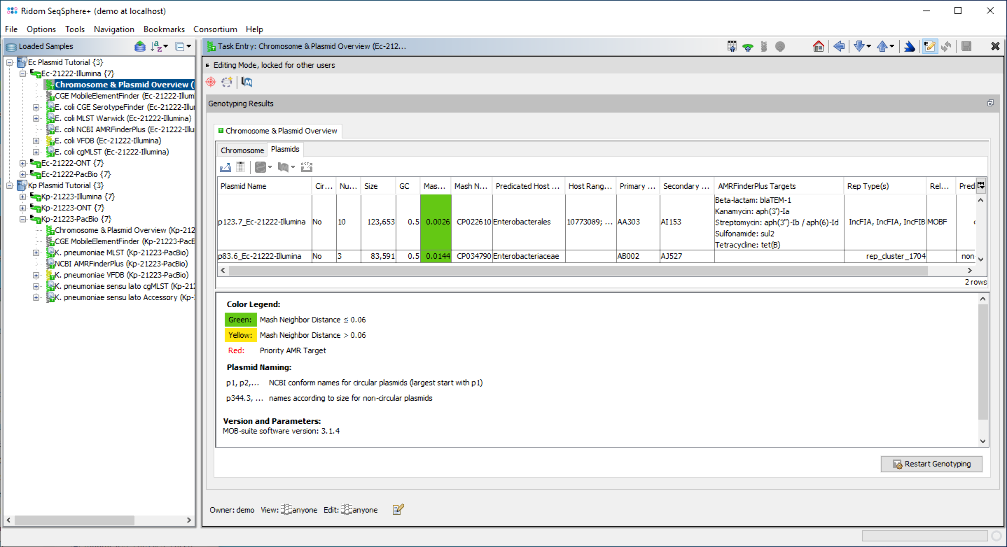
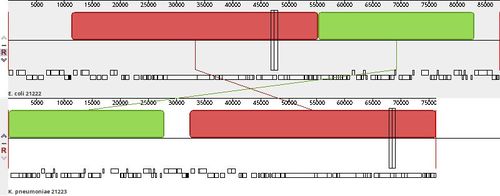
- For the Illumina data of Ec-21222-Illumina both reconstructed plasmids are non-circular and a blaCTX gene was not found at all in a plasmid or on the chromosome most likely due to the repetitive nature of the blaCTX-M-27 cassette. A plasmid with 84 kb also got a different Secondary Cluster ID (AJ527).
- For Kp-21223-Illumina a non-circular plasmid of size 63 kb containing blaCTX-M-27 was fairly well reconstructed with a Secondary Cluster ID AI281.
- Recovering Plasmids in the Absence of Long-Read Sequencing Data
- In 2021 Dutch microbiologist evaluated the performance of several plasmid reconstruction tools with short-read (Illumina) sequencing data and compared the results against the known finished plasmids of the isolates (Paganini et al. Microorganisms 9: 2021). MOB-suite/-recon showed the overall best performance. However, the authors concluded that even with this tool plasmid reconstruction from short-read data remains challenging. More specific the following problems were noted for MOB-recon:
- problems with small plasmids (plasmidSPAdes was performing here better),
- problems with plasmids carrying antibiotic resistance genes (all evaluated tools; due to an increased number of repetitive elements), and
- a tendency to split single plasmids into different predictions.
- Exactly the second problem is apparent from the Illumina data of the Ec-21222 isolate, whereas the third problem is illustrated by the reconstructed plasmid data of Kp-21223.
Further Analysis with Other Tools
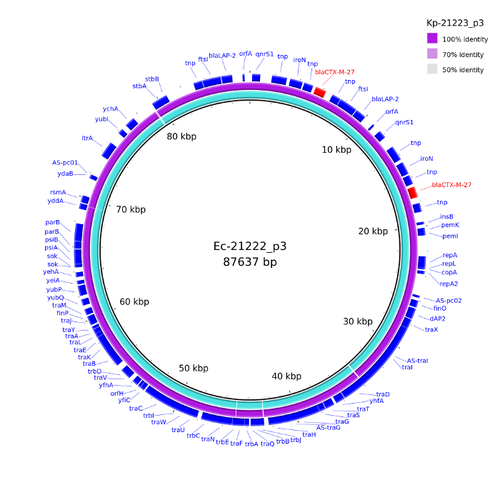
For doing further analysis and visualization of the two blaCTX-M-27 containing PacBio plasmids they can be exported to FASTA files. Open the Chromosome & Plasmid Overview, select in both PacBio Samples the third plasmid, and press the ![]() to export them to files.
to export them to files.
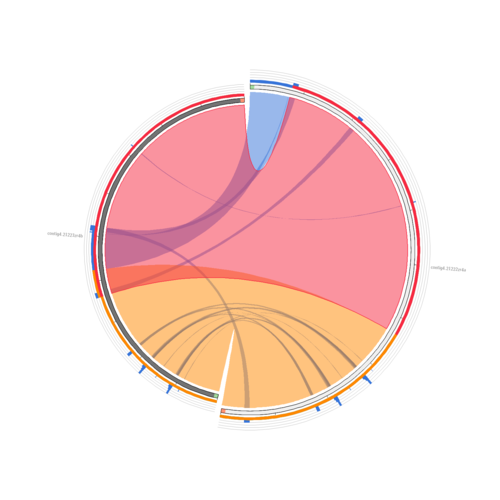
Compared with Circoletto