| wiki | search |
Contents |
Starting SeqSphere+
Be sure to install and initialize Ridom SeqSphere+ server and client.
After you are successfully logged in to a SeqSphere+ server, the welcome page is shown. It shows the server's welcome message, shortcuts for often used functions, the Samples opened in your previous session, and your bookmarks. The message and the shortcuts of the welcome page are configurable.
Basic Workflow in SeqSphere+
Define a Project
Ridom SeqSphere+ is designed for resequencing projects. You create and to set up a Project first. Each Project contains one or more Task Templates. A Task Template describes the targets that should be analyzed and the processing that should be performed, like automatic quality checks (e.g. minimum coverage) and genotyping (e.g. MLST).
For standardized and often used genotyping methods (like MLST or MLST+) predefined Task Templates can be easily downloaded from the Task Template Store. Customized Task Templates for other workflows can be created with a step-by-step dialog.
Import Sequence Data
There are basically two different kinds of sequence data that can be imported:
Sanger sequencing data (e.g., SCF or ABI chromatogram files) can be imported and automatically assembled. The sequence data is sorted into the defined targets by a configurable file naming convention.
Genome sequence data or contig libraries that were assembled from NGS reads (e.g., IonTorrent PGM) can be imported to extract the defined targets, including the aligned read data
All further processing and analysis is done on target level (i.e., gene-by-gene approach).
Quality Checks
The targets are automatically checked for the quality issues that were defined in the Task Template. To each target entry an analysis state (failed/succeeded) is assigned.
A list of all error positions in the imported data can be shown, and can be used to control those positions. Especially for PGM data, a reference based auto-correction of homopolymer related insertion/deletion errors is integrated.
Query Libraries
Each Task Template can have one or more Query Libraries to perform the typing or identification queries (e.g., MLST and MLST+).
After the queries of Task Entry has been performed, a result state assigned:
The results of a query (e.g., allele types) are stored in the result fields of the Task Entry.
Store in Database
Ridom SeqSphere+ consists of a client and a server software. The server can be accessed by multiple client from different locations, using different user accounts. The server contains an integrated database. This database stores the target based sequence data, the query library results and additional data like epidemiological information in a data entry called Sample.
Comparison Table
Finally the comparison table function can be used for advanced analysis of the typing results. The data can be processed and visualized using phylogenetic trees and minimum spanning trees.
Batch Mode and Pipeline Mode
Ridom SeqSphere+ is designed for resequencing projects. Therefore the import and processing steps, once established, are normally started as batch mode. The batch mode allows to choose a Project and a list of input files, to import, check, type and store multiple data entries in one continuous process.
Ridom SeqSphere+ also offers the non-interactive pipeline mode to assemble, import, type and analyze hundreds of your NGS sequence data at once automatically.
Data Objects in SeqSphere+
The Sample (![]() ) is the primary data object in SeqSphere+.
) is the primary data object in SeqSphere+.
Each Sample belongs to one Project (![]() ):
):
- Each Project has one Database Schema that specifies the Sample's database fields (e.g. sample ID, isolation date, etc.).
- Each Project has Task Templates that specify the Sample's DNA sequencing tasks (e.g. MLST).
Each Sample can have Task Entries (![]() ) that are specified by the Task Templates of its Project:
) that are specified by the Task Templates of its Project:
- Each Task Entry is meant for a specific DNA sequencing task that is performed for this Sample.
- Each Task Entry is specified by a Task Template that is available in the Sample's Project.
- Each Task Entry can have targets (
 ) that are specified in the Task Template.
) that are specified in the Task Template.
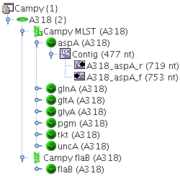
Example: A Project Campy, contains two Task Templates: Campy MLST, that contains the 7 MLST loci as targets, and Campy flaB that contains only a single target. Each Sample of this Project may now contain one Task Entry for Campy MLST, that contains up to 7 different targets, and one Task Entry for Campy flaB, that may contain only one target.