| wiki | search |
The Ridom MLVA Compare software allows easy analysis of MLVA, SNP and other categorical typing data (e.g. MLST results). Ridom MLVA Compare works with MS Excel and CSV files.
Plugins simplify working with MIRU-VNTR data (MLVA for Mycobacterium tuberculosis complex bacteria) and offer a quick connection to the MIRU-VNTRplus reference database for this kind of data.
Manual
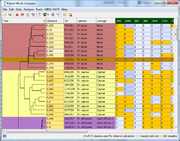 The Comparison Table displays all data in a tabular form. A tree can be included for ordering, and Samples groups can be visualized by color. | 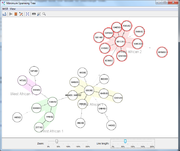 Minimum Spanning Tree (MST) can be calculated for the Samples. |
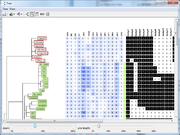 Phylogenetic trees can be calculated for the Samples. |  Different display options are available for the phylogenetic trees. |
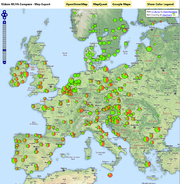
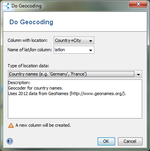 Geocoding can be done based on country names or Postal codes. | 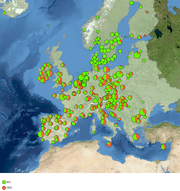 Maps can be created in a simple step for the geocoded Samples (Example: MRSA in Europe, Background image: Blue Marble from Nasa). |
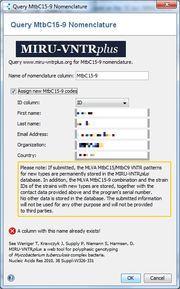 The MtbC15-9 nomenclature server can be queried. | |
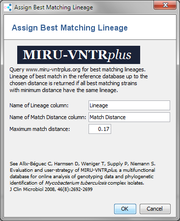 Simple MTBC Lineage identification using MIRU-VNTR data and MIRU-VNTRplus. |
- Working with the data
- Comparison Table
- Minimum Spanning Tree (MST)
- Phylogenetic trees
- Plugins
- Map Plugin
- MIRU-VNTR Plugin
- Appendix
- Preferences
Gene Mapper Import
Name Matching
Compare Table with data in file
Remove_Columns_With_Empty_Values
Map_Columns_For_Import
Latitude_and_longitude
Image File Formats
Set_Groups_by_Value